Details of DPV and References
DPV NO: 230 September 1980
Family: Tymoviridae
Genus: Tymovirus
Species: Turnip yellow mosaic virus | Acronym: TYMV
This is a revised version of DPV 2
Turnip yellow mosaic virus
R. E. F. Matthews Department of Cell Biology, University of Auckland, New Zealand
Contents
- Introduction
- Main Diseases
- Geographical Distribution
- Host Range and Symptomatology
- Strains
- Transmission by Vectors
- Transmission through Seed
- Transmission by Grafting
- Transmission by Dodder
- Serology
- Nucleic Acid Hybridization
- Relationships
- Stability in Sap
- Purification
- Properties of Particles
- Particle Structure
- Particle Composition
- Properties of Infective Nucleic Acid
- Molecular Structure
- Genome Properties
- Satellite
- Relations with Cells and Tissues
- Ecology and Control
- Notes
- Acknowledgements
- Figures
- References
Introduction
Described by Markham & Smith (1949).
A virus with RNA-containing icosahedral particles 28 nm in diameter. Host range is confined almost entirely to the Cruciferae. Infected cells show a characteristic rounding and clumping of the chloroplasts, readily observed by light microscopy. The virus may reach high concentrations in infected leaves. It is readily transmissible by sap inoculation, and is transmitted in the field by flea-beetles. It has been reported from several European countries.
Main Diseases
Causes mosaic disease in various Brassica spp.
Geographical Distribution
Europe.
Host Range and Symptomatology
Known host range is confined to Cruciferae (many species susceptible by mechanical inoculation) and one species in each of two related families: Reseda odorata in the Resedaceae and Cleome spinosa in the Capparidaceae.
-
Diagnostic species
- There is no definitive diagnostic species. Many strains give a bright yellow or yellow green mosaic in Brassica pekinensis (Chinese cabbage) but other strains give much milder symptoms (Fig. 1).
-
Propagation species
- Chinese cabbage is a good species for maintaining cultures, and for propagating the virus for purification. The best growing temperatures are in the range 20-25°C. Yields of virus are significantly reduced above 25°C.
-
Assay species
- There is no host that gives obvious local lesions under all conditions. Most virus strains give chlorotic local lesions in Chinese cabbage. These can be used for assay purposes in well nourished young plants grown at 18-22°C in moderate light intensity. Under other conditions, local lesions may be indistinct or may not form at all. Some strains, especially those isolated from white areas in the mosaic, produce necrotic local lesions. It is possible to obtain infection with 10-30 virus particles in 0.1-1.0 µl volumes of inoculum (Fraser & Matthews, 1979a). Chinese cabbage is also useful for experiments with beetle vectors.
Strains
The Cambridge culture (Markham & Smith, 1949). Stock cultures of the virus originating from Cambridge, England, consist of a mixture of closely related strains. Areas of different colour in leaves showing mosaic contain different strains of the virus (Fig. 1). It is not possible to isolate and maintain a pure culture of a single strain; single lesion isolates soon revert to a mixture like the original stock culture.
The Northumberland isolate (Broadbent & Heathcote, 1958). This isolate is serologically distinguishable from the Cambridge culture, and causes less severe symptoms in most hosts, but much more severe ones in cabbage, cauliflower and Brussels sprout.
Other strains collected in the field. Symons et al. (1963) studied the RNA and protein composition of six isolates obtained from various sources in England, Germany and Denmark. They fell clearly into two groups based on their cytosine contents (approximately 38%, or 42%). Analyses of the coat protein indicated a similar grouping.
Four strains were recognized among isolates collected in the German Democratic Republic (Shukla & Schmelzer, 1974). These could be distinguished both serologically and by symptoms on test plants.
Transmission by Vectors
The virus is transmitted in the field by species of Phyllotreta and Psylliodes (flea-beetles) (Markham & Smith, 1949). The mustard beetle (Phaedon cochleariae) and its larvae can also transmit. Larvae acquire the virus in 1-3 minutes’ feeding. There is a delay of about 1 day before they can infect plants. Ability to transmit is not retained through the pupal stage. Transmission is probably a purely mechanical process. It has not been established that regurgitation during feeding plays any role in natural transmission by undisturbed beetles.
Transmission through Seed
None recorded.
Transmission by Dodder
Not transmitted by Cuscuta sp. (Markham & Smith, 1949).
Serology
The nucleoprotein is strongly immunogenic in rabbits, the empty protein shells much less so. Most forms of serological test can be used for identification or assay of the virus. Preparations of the protein subunit of the virus shell do not cross-react in precipitation tests with antiserum prepared against the intact virus shell.
Relationships
Strains of the virus can be distinguished most readily by differences in the symptoms induced in Chinese cabbage and some other hosts, and by the severity of their effects on chloroplasts. Strains isolated by dissection of leaf tissue from a single plant (Fig. 1) appear serologically identical. Strains collected in the field may show serological differences detectable by immunodiffusion. The virus shows more or less distant serological relationships with many other tymoviruses. It is serologically unrelated to some of them (Koenig & Lesemann, 1980).
Stability in Sap
In Chinese cabbage sap at pH c. 6.0 some infectivity is retained after 10 min at 70°C but not at 75°C. Heat inactivation is very much more rapid at pH 8.5. At pH 6 to 7 infectivity decreases to zero over a period of weeks either at room temperature or at 4°C. Dilution end-point in sap from fully infected plants is usually between 10-4 and 10-6.
Purification
The virus is readily isolated on a large scale in good yield (0.5-2.0 mg/g fresh wt) from Chinese cabbage plants by the ethanol-ammonium sulphate method of Markham & Smith (1949), provided that the temperature is below 15°C while the virus is in contact with 30% ethanol. The pH 4.8 procedure of Matthews (1960) is convenient for preparation of virus from a few hundred ml or less of leaf extract. Clarification of larger volumes by adjustment to pH 4.8 and standing overnight is a safe alternative to the ethanol step of Markham & Smith (1949). The virus crystallizes as octahedra from solutions of ammonium sulphate (Fig. 2).
Properties of Particles
Purified virus preparations are resolved into numerous components when centrifuged in CsCl density gradients (Fig. 7). There is some confusion in terminology (cf. Mellema et al., 1979; Keeling, Collins & Matthews, 1979) but these two laboratories have now agreed to standardize on the teminology used here (C. W. A. Pleij, personal communication). The components are of three types:
(i) Empty protein shells, called T (for top component).
(ii) Infective nucleoproteins (B or bottom components) and particles derived from them. B component separates in CsCl gradients into components B1a and B1b, which are equally infective (Matthews, 1974), and a third component B1c, only recently characterized, which is more dense than B1b. B1b and B1c both contain copies of the coat protein mRNA as well as a molecule of genome RNA (C. W. A. Pleij, personal communication). B1 components can be converted in strong solutions of CsCl, especially at pH > 6.5, to a B2 series with higher densities, designated B2a, B2b and B2c. Their formation is prevented by the presence of 0.1 M MgCl2 in the CsCl (C. W. A. Pleij, personal communication).
(iii) Non-infective nucleoprotein particles containing subgenomic RNA species and having densities in CsCl intermediate between those of the T and B components. Matthews (1960) described three such components. Keeling et al. (1979), using an improved density gradient, resolved eight as illustrated in Fig. 7. These are numbered 1-8 in order of increasing RNA content. C. W. A. Pleij (personal communication) has now isolated two further components; the numbering of the components must therefore remain flexible.
The properties listed below for B1 were determined on a mixture of B1a, B1b and (presumably) B1c.
Sedimentation coefficients (s20,w) at infinite dilution: 53-54 (T); 116-117 (B1).
Molecular weights: 3.6 x 106 (T); 5.4 x 106 (B1).
Diffusion coefficients (D20 x 10-7 cm2 sec-1); 1.51 (T), 1.55 (B1).
Isoelectric points: pH 3 .75 (T and B1).
Partial specific volumes: 0.733 (T); 0.661 (B1).
Absorbances at 260 nm (1 mg/ml, 1 cm light path): 0.96 (T); 9.6 (B1).
A260/A280: 0.81 (T); 1.51 (B1).
Buoyant densities in CsCl (g/cm3): 1.28 (T); 1.395 (B1a) and 1.402 (B1b).
Some properties of the minor non-infective nucleoprotein fractions have been determined by
Mellema et al. (1979)
and
Keeling et al. (1979).
They contain the coat protein
mRNA and one or more of eight other subgenomic RNA species of discrete size (see Particle
Composition) which have not been firmly allocated to particular nucleoprotein species. If all
possible packaging combinations exist there may in fact be a very large number of particles of
slightly differing density. Thus the following data may be subject to revision; the first
figure is the effective buoyant density in CsCl (g/cm3), the second is the ratio
A260/A280,
the third and fourth (where available) are the calculated %
RNA and the approximate total weight of RNA in the particle (x 10-6).
Component 1:-
1.294; 1.135; 5%; (mainly coat protein gene).
Component 2:- 1.312; 1.310; 9%; 0.5.
Component 3:-
1.315; 1.286; 11%; 0.6.
Component 4:- 1.330; 1.325.
Component 5:- 1.340; 1.303; 17%; 0.9.
Component 6:- 1.345; 1.452.
Component 7:- 1.357; 1.504; 25%; 1.4.
Component 8:- 1.363; 1.507;
28%; 16.
The proportion of total minor nucleoproteins relative to the infective nucleoproteins
is about 5% on a particle number basis.
Particle Structure
The virus has icosahedral symmetry, and is 28-29 nm in diameter. It is built of 180 somewhat banana-shaped protein subunits clustered into 20 hexamers and 12 pentamers (Finch & Klug, 1966). The triangulation number T = 3. Electron micrographs of negatively stained virus show 32 morphological subunits (Fig. 3). The detailed arrangement of the RNA within the particle is not established, but some of it is closely associated with the protein subunits and some occupies the central space in the shell, and is in a fairly highly ordered state.
Among the icosahedral viruses, turnip yellow mosaic virus has a relatively stable structure. Protein-protein interactions are strong, as evidenced by the existence of the stable empty protein shells. However when the virus is taken to pH 11.6 in 1 M KCl the particle swells rapidly (radius increases by about 4%) and the RNA escapes, leaving an empty shell (Kaper, 1975; Keeling et al., 1979). Escape of the RNA is accompanied by a loss from the shell of an amount of protein equivalent to five protein subunits. The minor nucleoproteins do not swell or lose RNA or protein under the same conditions (J. Keeling & R. E. F. Matthews, unpublished data).
Particle Composition
Nucleic acid: RNA, single-stranded. The genome is one piece of RNA, with M. Wt c. 2.0 x 106, containing about 6400 nucleotides, and constituting about 35% by weight of the B1a nucleoprotein component. Molar percentages of nucleotides for the type strain: G17.2; A22.4; C38.3; U22.1. Sedimentation coefficient (s20,w) of RNA in 0.1 M KCl-phosphate buffer, pH 7.0: 28.4 ± 0.5 S. About 5% of the total RNA in purified virus preparations is a RNA species of M. Wt 0.25 x 106 which probably occurs in component B1b (Mellema et al., 1979). It also occurs in most of the minor nucleoprotein components together with any of eight other subgenomic RNA species, which have M. Wt of 0.34, 0.36, 0.39, 0.59, 0.71, 0.76, 0.96 and 1.30 x 106 (Higgins, Whitfeld & Matthews, 1978).
Protein: About 65% of the virus by weight. There is probably only one kind of protein in the icosahedral shell. This has a M. Wt of 20,133, and contains 189 amino acid residues of known sequence (Peter et al., 1972).
Other components: Polyamines, mainly spermidine, make up about 1% by weight of the virus (Beer & Kosuge, 1970). No lipid, carbohydrate or enzyme activities are found in the virus.
Genome Properties
Both the genome RNA and the 0.25 x 106 M. Wt RNA are capped at their 5' ends with the structure m7' G5ppp5' G - - - - -. The 3' ends consist of a tRNA-like sequence ending - - - - - -ApCpCpAOH which accepts valine (Pinck et al., 1970). The 0.25 x 106 M. Wt RNA contains the coat protein cistron which is also located on the genome RNA towards the 3' end. The base sequence of the 0.25 x 106 M. Wt RNA is known (Guilley & Briand, 1978). Excluding the cap there are 695 nucleotides. Proceeding from the 5' end there is an untranslated region of 19 nucleotides, then the coat protein gene of 567 nucleotides, and finally an untranslated region of 109 nucleotides. This contains the sequence which can form a tRNA-like structure with a valine anticodon-CAC- properly positioned (Briand et al., 1977).
In the wheat germ system for in vitro protein synthesis the genome RNA is translated to give two large polypeptides with M. Wt c. 180,000 and 150,000 but there is no product corresponding to coat protein. Such a product is, however, translated from the 0.25 x 106 M. Wt RNA; this therefore probably acts in vivo as a messenger for coat protein (Ricard et al., 1977; Pleij et al., 1977; Higgins et al., 1978). The other sub-genomic RNA species each give a polypeptide product in the wheat germ system that represents approximately their full coding capacity; they share a common 5' terminus which may be the same as that of the genome RNA (Mellema et al., 1979) and they probably form a family of ‘readthrough’ messenger RNAs for polypeptides required for virus synthesis in vivo. This idea is supported by the fact that infection of u.v.-irradiated protoplasts induces the synthesis of a series of polypeptides with sizes that correspond approximately to those expected from the in vitro studies (Y. Sugimura & R. E. F. Matthews, unpublished data).
Relations with Cells and Tissues
In a fully infected Chinese cabbage plant, virus is present in all tissues including the apical meristem region, but it reaches its highest concentration in the leaf lamina. Fully infected palisade cells isolated from infected leaves as protoplasts contain about 107 virus particles per cell. However, dark green islands in the mosaic contain little or no virus and are normal cytologically. In infected cells, the chloroplasts become rounded and clumped in a characteristic fashion that can be observed by light microscopy (Fig. 4). Observations on individual protoplasts show that this process takes 2-4 h, and is synchronous in individual cells (Fraser & Matthews, 1979b). In the light these rounded chloroplasts may subsequently develop large vacuoles, giving them a sickle-shaped appearance. Less commonly, the chloroplasts become angular in outline before clumping; in the light such chloroplasts may subsequently fragment to give many small pieces. The type of chloroplast alteration induced depends on some function of the viral genome (Fraser & Matthews, 1979b) but both types require active photosynthetic electron transport.
Electron microscopy reveals a variety of other tissue abnormalities which depend on strain of virus and on the time after infection (Ushiyama & Matthews, 1970). Many of these changes are secondary effects occurring after virus replication has begun. The most constant and characteristic cytological effect of infection is the development of small vesicles near the periphery of the chloroplasts. These vesicles are formed by an invagination of both of the chloroplast bilayer membranes. The vesicle necks are open to the cytoplasm (Fig. 5, Fig. 6). The double-stranded RNA found in infected tissue, and the virus-induced RNA-dependent RNA polymerase are associated with these vesicles, which are therefore a major site of virus RNA synthesis (Laflèche et al., 1972). They are the earliest consequence of infection detectable by electron microscopy (Hatta & Matthews, 1974) and their presence has been used as a marker of infection to establish a sequence of cytological changes.
Following the appearance of scattered peripheral vesicles, the chloroplasts become rounded. Clusters of closely spaced vesicles can be observed with endoplasmic reticulum lying over them in the cytoplasm (Fig. 8). The endoplasmic reticulum is then replaced by electron lucent material which is virus coat protein. Virus particles can be detected first in the cytoplasm overlying the electron lucent material (Fig. 9). Virus protein, probably in the form of empty protein shells, accumulates in the nuclei from an early stage after infection (Hatta & Matthews, 1976).
Brassica protoplasts can be infected in vitro. Virus accumulates between about 12 and 36 h after inoculation to give about 106 particles/protoplast (Renaudin et al., 1975). This is only about one tenth of the yield from palisade mesophyll cells in infected leaves. In infected leaves the ratio of ‘empty’ protein shells /‘full’ nucleoprotein particles is usually in the range 0.2 to 0.3. In protoplasts infected in vitro it is close to 1.0 throughout the virus growth cycle.
Notes
Several viruses may be confused with turnip yellow mosaic virus. Turnip mosaic virus (a potyvirus) may cause similar mosaic symptoms in turnip and Chinese cabbage. Turnip crinkle virus (a possible tombusvirus) and turnip rosette virus (a probable member of the southern bean mosaic virus group) are transmitted by flea beetles and have a stability in vitro similar to that of turnip yellow mosaic virus. Turnip rosette virus has a similar host range and causes rather similar symptoms. Erysimum latent virus is the only virus from cruciferous plants that resembles turnip yellow mosaic virus in showing two major sedimenting components (empty protein shells and virus nucleoprotein) on analytical centrifugation of leaf extracts, and in causing characteristic vesiculation of chloroplasts. These two viruses may be distinguished by serological tests.
Figures

Mosaic symptoms in Chinese cabbage. Top left is a healthy leaf. The other leaves are from plants inoculated with strains isolated from different islands of tissue in leaves containing the Cambridge stock culture. Top right, ‘pale green’; bottom left, ‘yellow green’; bottom right, ‘white’. (Photo. J. Endt.)

An octahedral crystal of virus particles grown in 0.75 M ammonium sulphate. Bar represents 25 µm. (Photo. T. Hatta.)
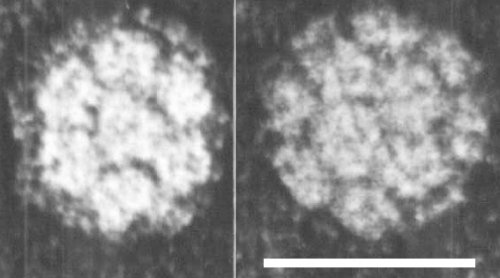
Morphological subunits seen in virus particles negatively stained with uranyl acetate. Bar represents 30 nm. (Photo. S. Bullivant.)
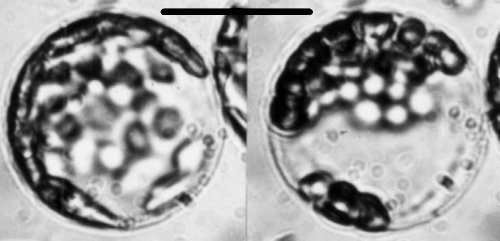
Development of rounding and clumping of chloroplasts in a Chinese cabbage mesophyll protoplast inoculated in vitro: (left) 12 h after inoculation, chloroplasts essentially normal; (right) the same protoplast 17 h after inoculation, rounding and clumping of chloroplasts complete. Bar represents 15 µm. (Photo. Y. Sugimura.)
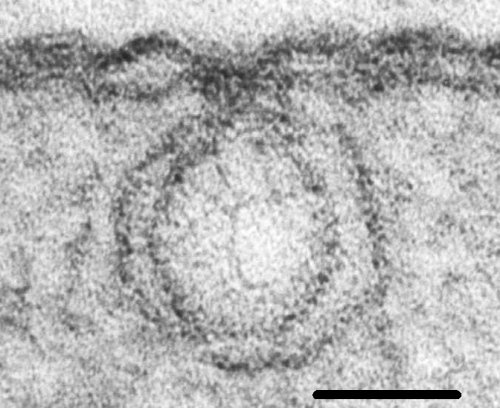
Small peripheral vesicles with necks, induced in Chinese cabbage chloroplasts. Thin section showing continuity of the vesicle membranes with the chloroplast membranes, and stranded material with the staining properties of double-stranded nucleic acid. Bar represents 50 nm. (Photo. S. Bullivant.)

Small peripheral vesicles with necks, induced in Chinese cabbage chloroplasts. Fine structure of vesicle membranes and necks revealed by freeze-fracturing. Bar represents 100 nm. (From Hatta, Bullivant & Matthews, 1973.)

Fractionation of a purified virus preparation into 12 nucleoprotein bands and a band of empty protein shells (T) in a CsCl gradient. Separation of the two infective nucleoprotein bands (B1a and B1b) is obscured by the overloading necessary to reveal some of the minor components (From Keeling et al., 1979.)

Chloroplast in an infected Chinese cabbage cell showing the clustered vesicles at the time they are overlaid by endoplasmic reticulum (ER) in the cytoplasm. Arrow shows a connection between ER and chloroplast membrane. Bar represents 250 nm. (From Hatta & Matthews, 1974.)

Chloroplast in an infected Chinese cabbage cell at a slightly later stage than that in Fig. 8. Overlying the electron-lucent material is a small array of virus particles. The tissue was plasmolysed with sucrose before fixation to induce crystallisation of any virus present. Bar represents 250 nm. (From Hatta & Matthews, 1974.)
References list for DPV: Turnip yellow mosaic virus (230)
- Beer & Kosuge, Virology 40: 930, 1970.
- Briand, Jonard, Guilley, Richards & Hirth, Eur. J. Biochem. 72: 453, 1977.
- Broadbent & Heathcote, Ann. appl. Biol. 46: 585, 1958.
- Finch & Klug, J. molec. Biol. 15: 344, 1966.
- Fraser & Matthews, J. gen. Virol. 44 : 565, 1979a.
- Fraser & Matthews, J. gen. Virol. 45: 623, 1979b.
- Guilley & Briand, Cell 15: 113, 1978.
- Hatta & Matthews, Virology 59: 383, 1974.
- Hatta & Matthews, Virology 73: 1, 1976.
- Hatta, Bullivant & Matthews, J. gen. Virol. 20: 37, 1973.
- Higgins, Whitfeld & Matthews, Virology 84: 153, 1978.
- Kaper, The Chemical Basis of Virus Structure, Dissociation and Reassembly, Amsterdam: North-Holland, New York: American Elsevier, 485 pp., 1975.
- Keeling, Collins & Matthews, Virology 97: 100, 1979.
- Koenig & Lesemann, CMI/AAB Descriptions of Plant Viruses 214, 5 pp., 1980.
- Laflèche, Bové, Dupont, Mouchés, Astier, Gamier & Bové, Proc. 8th FEBS Meeting, Amsterdam, 27: 43, Amsterdam: North Holland, 1972.
- Markham & Smith, Parasitology 39: 330, 1949.
- Matthews, Virology 12: 521, 1960.
- Matthews, Virology 60: 54, 1974.
- Mellema, Benicourt, Haenni, Noort, Pleij & Bosch, Virology 96: 38, 1979.
- Peter, Stehelin, Reinbolt, Collot & Duranton, Virology 49: 615, 1972.
- Pinck, Yot, Chapeville & Duranton, Nature, Lond. 226: 954, 1970.
- Pleij, Mellema, Noort & Bosch, FEBS Lett. 80: 19, 1977.
- Renaudin, Bové, Otsuki & Takebe, Molec. gen. Genet. 141: 59, 1975.
- Ricard, Barreau, Renaudin, Mouchés & Bové, Virology 79: 231, 1977.
- Shukla & Schmelzer, Acta phytopath Acad. Sci. Hung. 9: 237, 1974.
- Symons, Rees, Short & Markham, J. molec. Biol. 6: 1, 1963.
- Ushiyama & Matthews, Virology 42: 293, 1970.