Details of DPV and References
DPV NO: 366 September 1998
Family:
Genus:
Species: | Acronym:
This partially supercedes the Potyvirus Group description (245) which is mostly a Potyvirus genus description
Potyviridae family
D. D. Shukla CSIRO, Division of Molecular Science, Parkville, Victoria, Australia
C. W. Ward CSIRO, Division of Molecular Science, Parkville, Victoria, Australia
A. A. Brunt Horticulture Research International, Wellesbourne, Warwickshire, UK
P. H. Berger Department of Plant, Soil and Entomological Sciences, University of Idaho, Moscow, USA
Contents
- Type Member
- Main Characteristics
- Members
- Geographical Distribution
- Transmission by Vectors
- Ecology and Control
- Relations with Cells and Tissues
- Properties of Particles
- Genome Properties
- Replication
- Satellite
- Defective-Interfering RNA
- Relationships
- Notes on Tentative Members
- Affinities with Other Groups
- Notes
- Acknowledgements
- Figures
- References
Type Member
Potyvirus: Potato virus Y
Macluravirus: Maclura mosaic virus
Ipomovirus: Sweet potato mild mottle virus
Tritimovirus: Wheat streak mosaic virus
Rymovirus: Ryegrass mosaic virus
Bymovirus: Barley yellow mosaic virus
Main Characteristics
Flexuous filamentous particles, 11-15 nm in diameter with a helical pitch of c. 3.4 nm; those of genera with monopartite genomes (Potyvirus, Macluravirus, Ipomovirus, Tritimovirus, Rymovirus) are 650-950 nm long and those of the genus with bipartite genome (Bymovirus) are 200-300 and 500-600 long. Particles of monopartite genera members have a sedimentation coefficient (s20,w) of 150-160 and a density in CsCl of 1.31 g cm-3; those of bipartite members have a density in CsCl of 1.29 g cm-3. The particles consist of 95% coat protein and 5% RNA. The genome is single-stranded, positive-sense RNA. The genome size of monopartite members is c. 8.0 to 11 kb (mol. wt 2.7 x 106 to 3.0 x 106); the sizes of the two genome parts of bipartite members are 7.5 kb (mol. wt 2.6 x 103) and 3.5 kb (mol. wt 1.5 x 103). The genomic RNAs, like those of the picorna-like supergroup of viruses, have a genome-linked protein (VPg) of mol. wt c. 24,000 covalently bound to the 5' terminus and a poly(A) tail at the 3' end. Most members of the family Potyviridae have a coat protein of 30-37 kDa; that of WSMV (Tritimovirus) and MacMV (Macluravirus) is c. 40 kDa. All members of the Potyviridae induce the formation of cytoplasmic cylindrical inclusions (‘pinwheels’) consisting of a protein of c. 70 kDa.Some members also induce the formation of nuclear inclusions which consist of two proteins (c. 49 and 58 kDa) and/or amorphous proteinaceous inclusions (c. 56 kDa). Monopartite members are transmitted by aphids (Potyvirus, Macluravirus), mites (Rymovirus, Tritimovirus) or whiteflies (Ipomovirus), and bipartite members (Bymovirus) by the fungus Polymyxa graminis. Except for some bymoviruses, the viruses are transmissible by inoculation of sap; some potyviruses are carried within or on the testae of a small proportion of the seeds of some host species.
Most members of the Potyviridae have restricted, or very restricted, host ranges, but a few occur naturally in a wide range of monocotyledonous and/or dicotyledonous species. However, members of the Potyviridae infect many important crop species. Some induce no conspicuous symptoms in infected plants, but most cause mosaic or mottle symptoms in leaves; many also induce colour-breaking in flowers, mottled and/or distorted fruits and seeds, and some cause considerable losses of crop yield and quality.
Members
Definitive and possible members of each of the six genera with their acronyms are listed below:
Table 1. Members and possible members of the six genera of the
Potyviridae*
| Virus or species*** | Acronym** |
| Genus: Potyvirus | |
| Definitive Species: | |
| Alstroemeria mosaic virus | AlMV |
| Amaranthus leaf mottle virus | AmLMV |
| Araujia mosaic virus | ArjMV |
| artichoke latent virus | ALV |
| asparagus virus 1 | AV-1 |
| bean common mosaic virus (73, 337) | BCMV |
| (Azuki bean mosaic virus) | |
| (blackeye cowpea mosaic virus) (305) | |
| (Dendrobium mosaic virus) | |
| (guar green sterile virus) | |
| (peanut stripe virus) | |
| (peanut mild mottle virus) | |
| (peanut chlorotic ring mottle virus) | |
| (some cowpea aphid-borne mosaic virus strains) | |
| bean common mosaic necrosis virus | BCMNV |
| (serotype A of BCMV) | |
| bean yellow mosaic virus (40) | BYMV |
| (Crocus tomasinianus virus) | |
| (white lupin mosaic virus) | |
| (pea mosaic virus) | |
| beet mosaic virus (53) | BtMV |
| bidens mottle virus (161) | BiMoV |
| calanthe mild mosaic virus | CalMMV |
| cardamom mosaic virus | CdMV |
| carnation vein mottle virus (78) | CVMoV |
| carrot thin leaf virus (218) | CTLV |
| celery mosaic virus (50) | CeMV |
| chilli veinal mottle virus | ChiVMV |
| (pepper vein banding mosaic virus) | |
| clover yellow vein virus (131) | ClYVV |
| (pea necrosis virus) | |
| (statice virus Y) | |
| cocksfoot streak virus (59) | CSV |
| Colombian datura virus | CDV |
| Commelina mosaic virus | ComMV |
| Cowpea aphid-borne mosaic virus (134) | CABMV |
| (South African passiflora virus) | |
| cowpea green vein banding virus | CGVBV |
| dasheen mosaic virus (191) | DsMV |
| datura shoestring virus | DSTV |
| endive necrotic mosaic virus | ENMV |
| freesia mosaic virus | FreMV |
| Gloriosa stripe mosaic virus | GSMV |
| groundnut eyespot virus | GEV |
| guinea grass mosaic virus (190) | GGMV |
| Helenium virus Y | HVY |
| henbane mosaic virus (95) | HMV |
| Hippeastrum mosaic virus (117) | HiMV |
| hyacinth mosaic virus | HyaMV |
| iris fulva mosaic virus (310) | IFMV |
| iris mild mosaic virus (116, 324) | IMMV |
| iris severe mosaic virus (338) | ISMV |
| (bearded iris mosaic virus) (147) | |
| Johnsongrass mosaic virus (340) | JGMV |
| Kalanchoe mosaic virus (Husted et al., 1994) | KMV |
| konjak mosaic virus | KonMV |
| leek yellow stripe virus (240) | LYSV |
| (garlic potyvirus) | |
| (garlic virus 2) | |
| lettuce mosaic virus (9) | LMV |
| lily mottle virus | LiMoV |
| maize dwarf mosaic virus (341) | MDMV |
| Moroccan watermelon mosaic virus | WMMV |
| narcissus degeneration virus | NDV |
| narcissus late season yellows virus (367) | NLSYV |
| (jonquil mild mosaic virus) | |
| narcissus yellow stripe virus (76) | NYSV |
| nerine yellow stripe virus | NeYSV |
| Nothoscordum mosaic virus | NoMV |
| onion yellow dwarf (158) | OYDV |
| Ornithogalum mosaic virus | OrMV |
| papaya ringspot virus (84, 292) | PRSV |
| (watermelon mosaic virus 1) (63) | |
| parsnip mosaic virus (91) | ParMV |
| passionfruit woodiness virus (122) | PWV |
| pea seed-borne mosaic virus (146) | PSbMV |
| peanut mottle virus (141) | PeMoV |
| pepper mottle virus (253) | PepMoV |
| pepper severe mosaic virus | PeSMV |
| pepper veinal mottle virus (104) | PVMV |
| Peru tomato virus (255) | PTV |
| petunia flower mottle virus | PFMoV |
| plum pox virus (70) | PPV |
| pokeweed mosaic virus (97) | PkMV |
| potato virus A (54) | PVA |
| potato virus V (316) | PVV |
| potato virus Y (37, 242) | PVY |
| Rembrandt tulip breaking virus | RTBV |
| sesame mosaic virus | SeMV |
| shallot yellow stripe virus | SYSV |
| (Welsh onion yellow stripe virus) | |
| sorghum mosaic virus (359) | SrMV |
| soybean mosaic virus (93) | SMV |
| sugarcane mosaic virus (88, 342) | SCMV |
| sweet potato feathery mottle virus | SPFMV |
| (sweet potato russet crack) | |
| (sweet potato A) | |
| (sweet potato chlorotic leaf spot) | |
| (sweet potato internal cork) | |
| sweet potato latent virus | SwPLV |
| tamarillo mosaic virus | TamMV |
| Telfairia mosaic virus | TeMV |
| tobacco etch virus (55, 258) | TEV |
| tobacco vein banding mosaic virus | TVBMV |
| tobacco vein mottling virus (325) | TVMV |
| tulip breaking virus (71) | TBV |
| turnip mosaic virus (8) | TuMV |
| (tulip top breaking virus) | |
| (tulip chlorotic blotch virus) | |
| Wakegi yellow dwarf virus (Tsuneyoshi et al., 1998) | WYDV |
| watermelon mosaic virus 2 (63, 293) | WMV-2 |
| (vanilla necrosis virus) | |
| Wisteria vein mosaic virus | WVMV |
| yam mild mosaic virus (Mumford & Seal, 1997) | YMMV |
| yam mosaic virus (314) | YMV |
| (Dioscorea green banding virus) | |
| zucchini yellow fleck virus | ZYFV |
| zucchini yellow mosaic virus (282) | ZYMV |
| Possible Species: | |
| Amazon lily mosaic virus | AliMV |
| Aneilema virusb | AneV |
| Anthoxanthum mosaic virusa | AntMV |
| Aquilegia virusa,b | AqV |
| Arracacha virus Y | AVY |
| Asystasia gangetica mottle virusa | AGMoV |
| bidens mosaic virus | BiMV |
| bramble yellow mosaic virus | BrmYMV |
| Bryonia mottle virus | BryMoV |
| Calanthe mosaic virus | CalMV |
| canary reed mosaic virus | CRMV |
| Canavalia maritima mosaic virus | CnMMV |
| carrot mosaic virus | CtMV |
| Cassia yellow spot virus | CasYSP |
| celery yellow mosaic virus | CeYMV |
| chickpea bushy dwarf virus | CpBDV |
| chickpea filiform virus | CpFV |
| clitoria yellow mosaic virus | CtYMV |
| cowpea rugose mosaic virus | CPRMV |
| Crinum mosaic virusa | CriMV |
| Croatian clover virusb | CroCV |
| Cypripedium calceolus virusa | CypCV |
| daphne virus Y | DVY |
| datura virus 437 | DV-437 |
| datura distortion mosaic virus | DDMV |
| datura mosaic virusa | DTMV |
| datura necrosis virus | DNV |
| Desmodium mosaic virus | DesMV |
| Dioscorea trifida virusb | DTV |
| Dipladenia mosaic virus | DipMV |
| dock mottling mosaic virus | DMMV |
| eggplant green mosaic virus | EGMV |
| eggplant severe mottle virus | ESMoV |
| Euphorbia ringspot virusa | EuRV |
| Ficus carica virusb | FicCV |
| guar symptomless virusa | GSLV |
| Habenaria mosaic virus | HaMV |
| Holcus streak virusa | HSV |
| Hungarian Datura innoxia virusa | HDIV |
| Indian pepper mottle virus | IPMoV |
| isachne mosaic virusa | IsaMV |
| Kennedya virus Y | KVY |
| lily mild mottle virus | LiMMoV |
| Malva vein clearing virus | MVCV |
| marigold mottle virus | MaMoV |
| Melilotus mosaic virus | MeMV |
| Melon vein-banding mosaic virus | MVBMV |
| mungbean mosaic virusa | MbMV |
| mungbean mottle virus | MbMoV |
| Nasturtium mosaic virus | NasMV |
| nerine virus Y | NVY |
| palm mosaic virusa | PalMV |
| papaya leaf distortion mosaic virus | PLDMV |
| passionfruit mottle virus | PFMoV |
| passionfruit ringspot virus | PFRSV |
| patchouli mottle virus | PatMoV |
| peanut green mottle virus | PeGMoV |
| Pecteilis mosaic virus | PcMV |
| pepper mild mosaic virus | PMMV |
| pepper vein banding virus | PVBV |
| perilla mottle virus | PerMoV |
| plantain virus 7 | PlV-7 |
| Pleioblastus mosaic virus | PleMV |
| Populus virusa | PV |
| primula mosaic virus | PrMV |
| primula mottle virus | PrMoV |
| ranunculus mottle virus | RanMoV |
| rudbeckia mosaic virus | RuMV |
| Sri Lankan passionfruit mottle virus | SLPMoV |
| sunflower mosaic virusa | SuMV |
| sweet potato vein mosaic virus | SPVMV |
| sweet potato mild speckling virus | SPMSV |
| sword bean distortion mosaic virus | SBDMV |
| teasel mosaic virus | TeaMV |
| tobacco wilt virus | TWV |
| Tongan vanilla virus | TVV |
| Tradescantia/Zebrina virusb | TZV |
| Trichosanthes mottle virus | TrMoV |
| Tropaeolum virus 1 | TV-1 |
| Tropaeolum virus 2 | TV-2 |
| tulip band breaking virus | TBBV |
| Ullucus mosaic virus | UMV |
| Vallota mosaic virus | ValMV |
| vanilla mosaic virus | VanMV |
| white bryony virus | WBV |
| wild potato mosaic virus | WPMV |
| Zoysia mosaic virus | ZMV |
| Genus: Macluravirus | |
| Definitive Species: | |
| Maclura mosaic virus (239) | MacMV |
| Narcissus latent virus (170) | NLV |
| Possible Species: | |
| None reported | |
| Genus: Ipomovirus | |
| Definitive Species: | |
| sweet potato mild mottle virus (162) | SPMMV |
| Possible Species: | |
| sweet potato yellow dwarf virus | SPYDV |
| Genus: Tritimovirus | |
| Definitive Species: | |
| wheat streak mosaic virus (48) | WSMV |
| brome streak mosaic virus | BrSMV |
| Possible species: | |
| Sugarcane streak mosaic virus | SCSMV |
| Genus: Rymovirus | |
| Definitive Species: | |
| Agropyron mosaic virus (118) | AgMV |
| Hordeum mosaic virus | HoMV |
| oat necrotic mottle virus (169) | ONMV |
| ryegrass mosaic virus (86) | RGMV |
| Possible species: | |
| Spartina mottle virus | SpMV |
| Genus: Bymovirus | |
| Definitive species: | |
| barley yellow mosaic virus (143) | BaYMV |
| barley mild mosaic virus (356) | BaMMV |
| oat mosaic virus (145) | OMV |
| rice necrosis mosaic virus (172) | RNMV |
| wheat spindle streak mosaic virus (167) | WSSMV |
| wheat yellow mosaic virus (Namba et al., 1998) | WYMV |
| Possible species: | |
| None reported |
| * | As listed in Francki et al. (1991), and with amendments suggested by the ICTV Potyvirus Study Group (Berger et al., 1998). |
| ** | Acronyms of members as suggested by Hull et al. (1991) and Berger et al. (1998). |
| *** | Key References for each virus are listed in Shukla et al. (1994) and Berger et al. (1997). Numbers in parentheses are those of the relevant Description, if available. |
| a | Aphid transmission not confirmed. |
| b | Name inadequate, but denotes the species in which the virus was first found. |
Geographical Distribution
Potyviruses occur world-wide but are especially prevalent in tropical and subtropical countries. Rymoviruses have been reported in temperate regions of North America and Europe. Bymoviruses and tritimoviruses occur in temperate regions of North America, Europe and Japan. By contrast, ipomoviruses occur naturally only in tropical East Africa or Asia and macluraviruses have been reported only from Europe, USA and Australia (NLV) and the former Yugoslavia (MacMV).
Transmission by Vectors
The natural mode of vector transmission of members of the six genera is as
follows:
Potyvirus
Usually transmitted by aphids in a non-persistent, non-circulative manner.
Virus is acquired and transmitted most efficiently after brief feeding probes (a
few min); acquisition is enhanced if the aphids are previously starved for 1 to 3 h.
Aphids usually remain viruliferous for less than 1 h with continued feeding, or 4 h
if starved, although retention is much longer (up to 24 h) under some conditions.
There is no latent period between acquisition and transmission, and no virus
multiplication within the vector. Although some members are transmissible by
several or many aphid species, considerable virus-vector specificity occurs; there
are also marked differences in efficiency of transmission between different
clones and colonies of one aphid species, or between different virus
strains.
Potyvirus species tested so far require a ‘helper factor’ for their
transmission by aphids
(Govier & Kassanis, 1974);
this factor, designated
HC-Pro, is a labile protein of c. 56 kDa which is encoded in the viral
genome (Fig. 1)
and possibly facilitates the attachment of virus particles to the
mouth parts of the vector aphids
(Govier et al., 1977;
Pirone & Thornbury, 1983;
Thornbury et al., 1990).
A conserved three amino acid
motif, Asp-Ala-Gly (DAG), located in the surface-exposed, amino-terminal
domain of the coat protein also is involved in aphid transmission of
Potyvirus species
(Atreya et al., 1990,
1991;
Gal-On et al., 1992).
HC-Pro and virions have been shown to co-localize in the maxillary food
canal and foregut of aphids
(Berger & Pirone, 1986;
Ammar et al., 1994;
Wang et al., 1996),
and a direct interaction between HC-Pro and
coat protein of TVMV was demonstrated recently
(Blanc et al., 1997).
A
seven amino acid residue region (DTVDAGK), located in the N-terminus of the
coat protein and which includes the DAG motif, was shown to mediate HC-Pro
binding
(Blanc et al., 1997).
Thus, it appears that the long-standing
hypothesis
(Govier & Kassanis, 1974),
termed recently as the ‘bridge’
hypothesis by
Pirone & Blanc (1996),
to explain the aphid transmission
mechanism for potyviruses has been supported. According to this hypothesis
HC-Pro acts as a bifunctional molecule, one domain of which binds to the coat
protein and the other to putative receptor in the vector mouth parts.
Members of a few other invertebrate taxa have been reported to transmit
Potyvirus species: PRSV-W and CeMV were reported to be inefficiently
transmitted by the leaf miner Liriomyza sativae (Diptera: Agromyzidae)
(Zitter & Tsai, 1977);
and PSbMV and PVY by red spider mites
(Tetranychus urticae)
(Khetarpal et al., 1989). Apparent soil
transmission of SCMV in the absence of root contact suggested the involvement
of a soil-living vector
(Bond & Pirone, 1970).
SCMV-MDB was reported to
be transmitted in South Africa by the uredospores of Puccinia sp.
(Von Wechmar et al., 1992).
Macluravirus
Like species of the genus Potyvirus, are
transmitted by aphids in a non-persistent, stylet-borne manner
(Brunt, 1976,
1977;
Plese & Stefanac, 1976;
Koenig & Plese, 1981).
Ipomovirus
Transmitted by the whitefly Bemisia tabaci, possibly in a non-persistent
manner
(Hollings et al., 1976;
Liao et al., 1979).
Tritimovirus
Transmitted in a persistent manner by eriophyid mites (Abacarus hystrix
and Aceria tulipae)
(Shepard & Carroll, 1967;
Slykhuis, 1973;
Plumb & Jones, 1973;
Paliwal, 1980).
Rymovirus
Transmitted in a persistent-circulative manner by eriophyid mites (Abacarus hystrix
and Aceria tulipae)
(Shepard & Carroll, 1967;
Slykhuis, 1973;
Plumb & Jones, 1973;
Paliwal, 1980).
Bymovirus
Transmitted within zoospores of the chytrid fungus Polymyxa graminis
(Saito et al., 1968;
Inouye & Fujii, 1977;
Inouye & Saito, 1975),
and survives for many years within resting spores.
Ecology and Control
Potyvirus species, transmitted by aphids, are very successful
pathogens in a wide range of crops and environmental conditions; they are most
prevalent in tropical or subtropical countries, where continuous or successive
cropping is usually practised. Of the many different factors that affect their rate
of spread and severity in crops, the most important are the proximity and
prevalence of virus sources, and the number, activity and occurrence of vector
species. Early infection of the crop often results in extensive spread and thus a
high incidence and severe disease. In temperate climates, potyviruses survive in
perennial or vegetatively propagated crops but, because most have narrow,
often extremely restricted, host ranges, comparatively few naturally infect
alternative host species; even where this does occur, it is often of only minor
importance. The most important virus sources are infected seed, planting
material, or infected volunteer plants from previous crops
(Hollings & Brunt, 1981a,
1981b).
Host-specialization of different members has resulted in the occurrence of
numerous strains, or pathotypes, which may have survived through the absence
of inter-strain competition in common hosts
(Hollings & Brunt, 1981a,
1981b);
nevertheless, complexes of two or more different potyviruses are often
found in individual plants of some species.
The ecology of Macluravirus species has yet to be investigated,
but is probably similar to that of potyviruses
(Brunt, 1977;
Koenig & Plese, 1981).
Ipomovirus species are disseminated in infected propagation stock of
sweet potato and spread locally by their whitefly vector
(Hollings et al., 1976,
Liao et al., 1979).
Tritimovirus and Rymovirus species
and their vectors persist in wheat, maize, millets and susceptible grasses.
Spread is facilitated by overlapping summer and autumn-sown crops
(Slykhuis et al., 1957;
Atkinson & Slykhuis, 1963)
and by wind dispersal of
viruliferous mites.
Bymovirus species are disseminated within fungal resting spores by wind, water and farm equipment and the movement of soil during cultivation (Toler & Hebert, 1964; Slykhuis, 1970; Kusaba et al., 1971). Local spread by viruliferous zoospores is most rapid when soil moisture content is high.
Relations with Cells and Tissues
Virus particles are sometimes found scattered randomly throughout the
cytoplasm of infected cells, within plasmodesmata, in uniseriate arrays parallel to
the tonoplast or cytoplasmic lamellae, and/or in cytoplasmic strands projecting
into or bridging the cell vacuole (e.g.
Lawson & Hearon, 1971;
Weintraub et al., 1974).
Membrane-associated particle aggregates present in
negatively stained tissue extracts are possibly either fragments of the
virus-containing cytoplasmic strands or tonoplast-associated particles entrapped
during cellular disruption by pieces of the adjacent membrane
(Brunt & Atkey, 1974).
Infected cells sometimes contain unusual aggregates of
mitochondria or chloroplasts
(Kitajima & Lovisolo, 1972;
Kitajima & Costa, 1973;
Weintraub et al., 1973).
Cytoplasmic inclusions
Members of the family Potyviridae characteristically induce the formation of
cytoplasmic inclusions (CI), which are detectable by light microscopy in suitably
stained epidermal leaf cells
(Christie & Edwardson, 1977).
Their structure
has been elucidated from ultrastructural studies of infected plants
(Edwardson, 1966;
Rubio-Huertos & Lopez-Abella, 1966)
and freeze-etch electron
microscopy
(McDonald & Hiebert, 1974).
Inclusions each have a central
core from which radiate 10-20 thin striated rectangular or triangular curved plates
or ‘lamellae’, which have a surface lattice of 5 nm.
In transverse sections, CI are
seen as ‘pinwheels’ and in longitudinal sections as ‘bundles’.
Sections of
‘lamellae’, when not obviously associated with the ‘pinwheel’
core, are described
as ‘laminated inclusions’ if straight or unfolded,
as ‘scrolls’ or ‘tubes’ if rolled
inwardly, and as ‘laminated aggregates’ if two or more are closely associated or
partially fused. Morphologically, CI are generally considered to be either
cylindrical
(Edwardson, 1966)
or conical
(Andrews & Shalla, 1974).
Members have been subdivided into four sub-divisions on the basis of
morphology of their CI
(Edwardson, 1974;
Edwardson et al., 1984).
Viruses in the genera Macluravirus, Ipomovirus, Tritimovirus, Rymovirus and
Bymovirus, induce the formation of CI which are usually indistinguishable
from those of Potyvirus species
(Langenberg & Schroeder, 1973,
1974;
Plumb & Jones, 1973;
Hollings et al., 1976;
Plese & Stefanac, 1976;
Chamberlain & Catherall, 1977).
The CI of WSMV are
elliptic hyperboloids
(Mernaugh et al., 1980).
Viruses in the genus
Bymovirus induce massive accumulation of membranes and CI with
laminated aggregates but no scrolls
(Saito et al., 1966).
The WSMV
infected cells also contain hypertrophied endoplasmic reticulum, small vesicles
and abnormally numerous ribosomes
(Huth et al., 1984).
The lamellae and/or their fragments can be extracted and purified from infected
plants with yields (16-18 A280 units/kg leaf tissue)
sometimes considerably greater than those of virus particles
(Hiebert & McDonald, 1973).
Cytoplasmic inclusions contain a single virus-specific protein
of 67-70 kDa, identified as an RNA helicase
(Lain et al., 1990,
1991) (see
Fig. 1).
Nuclear inclusions
A few Potyvirus species, notably TEV, also induce the formation of
virus-encoded crystalline nuclear inclusions (NI) which, like CI, are readily detectable
by light microscopy in suitably stained epidermal cells
(Kassanis, 1939;
Christie & Edwardson, 1977).
The NI are flat crystals 6 to 8 µm square when
viewed from above, but are slightly curved plates in side view.
The NI of TEV can be extracted and purified, yields of 2 x 108 to
8 x 108 inclusions/kg leaf tissue being readily obtained
(Knuhtsen et al., 1974).
NI consist of thin rectangular plates which may be present
in regular stacks but are often offset at 45° to each other. The plates have
a clearly discernible lattice with striations 10.2 nm apart and having primary axes
intersecting at 90°
(McDonald & Hiebert, 1974). In transverse section,
the NI appear to be multilayered pyramids.
The NI of TEV contain two proteins designated NIa and NIb (49.8 and 54.5 kDa,
respectively) which are serologically unrelated to the capsid protein of the
associated virus particles, or to the CI protein or to healthy plant protein
(Knuhtsen et al., 1974).
The NIa codes for VPg-proteinase, while NIb is
the RNA-dependent RNA polymerase (Fig. 1).
NI of various shapes and sizes
induced by other potyviruses have yet to be similarly characterized
(Christie & Edwardson, 1977).
Amorphous inclusions
Some members of the Potyviridae also induce the formation of amorphous
inclusions (AI), which are aggregates of a single protein species (53-58 kDa), the
virus-encoded aphid transmission helper factor
(De Mejia et al., 1985a,
1985b;
Hellmann et al., 1988;
Baunoch et al., 1990).
Properties of Particles
Most members of the Potyviridae have flexuous filamentous particles 11
to 12 nm wide, made up of 1700-2000 copies of a single coat protein subunit
arranged in a helical manner around a single molecule of viral RNA.
The particles of monopartite genera have modal length of 650-675 nm
(Macluravirus), 680 to 750 nm (Tritimovirus), 690 to 720 nm
(Rymovirus), 720 to 850 nm
(Potyvirus) or 750 to 950 nm (Ipomovirus)
(Hollings & Brunt, 1981a,
1981b;
Koenig & Plese, 1981),
and those of the bipartite
genus (Bymovirus) have biomodal lengths of 200 to 300 nm and 500 to
600 nm
(Usugi et al., 1989).
Particles of some Potyvirus species
(such as HMV and PVMV) are straight and c. 850 nm long in the
presence of Mg++ ions, but otherwise flexuous and c. 750
nm long
(Govier & Woods, 1971).
Particles of members of the Potyviridae show very little substructure,
although some have a discernible central canal 2 to 3 nm diameter in negatively
stained preparations. The protein subunits have a helical pitch of c. 3 to
4 nm
(Varma et al., 1968).
A particle weight of 60 x 106 -
70 x 106 daltons, estimated from nucleic acid and protein
contents, indicates that each particle contains 1700-2000 protein subunits,
possibly arranged 7-8 per turn of the helix and with c. 6 nucleotides per
protein subunit
(Veerisetty, 1979).
Dissembled PVY coat protein readily forms
long (up to several µm) flexuous particles of non-helical stacked discs
(repeat distance 4 nm) in the absence of RNA
(Goodman et al., 1976;
McDonald et al., 1976;
McDonald & Bancroft, 1977). A schematic
diagram illustrating the tertiary structure of a virus particle of a typical member of
the Potyviridae is shown in Fig. 2.
It is based on the demonstrated
surface location of the N- and C-termini of the coat protein and the predictions of
secondary structure that suggest four major antiparallel alpha-helical segments
(Shukla et al., 1988;
Shukla & Ward, 1989a).
Members have U.V. absorbance spectra typical of nucleoproteins, with a
maximum at 260 to 262 nm and a minimum at 240 to 246 nm. The
A260/A280 ratios of 1.14-1.25, and
Amax/Amin ratios of 1.11-1.27 are
typical of nucleoproteins containing 5-6% nucleic acid
(Layne, 1957).
The
extinction coefficient (A0.1%, 1cm, 260nm), which has been
determined for only a few members, ranges from 2.4 to 2.9. Several members
are stable in CsCl at 20 to 25°C, and have buoyant densities of 1.318 to
1.336 g cm-3
(Huttinga & Mosch, 1974;
Damirdagh & Shepherd, 1970).
Coat protein weights have been estimated by sodium dodecyl sulphate-
polyacrylamide gel electrophoresis (SDS-PAGE), amino acid analysis, and
protein and gene sequencing. The values obtained by SDS-PAGE and amino
acid analysis have varied from the definitive values calculated from the coat
protein amino acid sequences (see
Shukla et al., 1994).
The coat protein
sequences of members of the Potyviridae show that the coat proteins
range in size from 253 to 332 amino acids
(Ward & Shukla, 1991;
Shukla et al., 1994;
Berger et al., 1997).
The coat proteins of members of the Potyviridae are often reported to be heterogeneous with two (Hiebert & McDonald, 1973; Huttinga & Mosch, 1974; Moghal & Francki, 1976), three (Choi & Wakimoto, 1979) or more components (Brakke et al., 1990; Niblett et al., 1991). Multiple bands (Huttinga & Mosch, 1974; Moghal & Francki, 1976) are attributed to partial degradation of the polypeptide by proteolytic enzymes of host or microbial origin, probably by removal of the N and C termini of the coat proteins (Shukla et al., 1988). A second explanation, at least for PRSV, is differential polyprotein processing at two or more sites in the junction between NIb and coat protein (Yeh et al., 1992).
Genome Properties
Members of the Potyviridae have particles containing 5 to 6% single-stranded
positive-sense RNA
(Matthews, 1979).
The genomes of the genera
Potyvirus, Macluravirus, Ipomovirus , Rymovirus and Tritimovirus
consist of one RNA molecule, estimated by PAGE for some Potyvirus
species to have Mr (x 10-6) of 3.5 (TuMV), 3.2 (PVY) and 3.15
(TEV)
(Hill & Shepherd, 1972;
Hinostroza-Orihuela, 1975;
Hari et al., 1979).
By density gradient centrifugation, the mol. wt of RNA of SCMV-MDB
was estimated to be 2.7 x 106 and that of PVY-RNA to be 3.1 x
106
(Pring & Langenberg, 1972;
Makkouk & Gumpf, 1974,
1975).
Complete genome sequences are now available for at least 20
species of the genus Potyvirus, one species of the genus
Ipomovirus and two species each of the genera Tritimovirus
and Bymovirus.
The Potyvirus species for which the complete nucleotide sequences
presently available are: TEV
(Allison et al., 1986;
Chu et al., 1995),
TVMV
(Domier et al., 1986;
Nicolas et al., 1996),
PPV
(Lain et al., 1989;
Maiss et al., 1989;
Teycheney et al., 1989;
Palcovics et al., 1993),
PVY
(Robaglia et al., 1989;
Thole et al., 1993;
Singh & Singh, 1996;
Jakab et al., 1997),
PSbMV
(Johansen et al., 1991,
1996),
SMV
(Jayaram et al., 1992),
TuMV
(Nicolas & Laliberte, 1992;
Oshima et al., 1996),
PepMoV
(Vance et al., 1992),
PRSV
(Yeh et al., 1992),
JGMV
(Gough & Shukla, 1993),
PStV
(Gunasinghe et al., 1994),
PVA
(Puurand et al., 1994),
BCMNV
(Fang et al., 1995),
ZYMV
(Wisler et al., 1995),
YMV
(Aleman et al., 1996),
BYMV
(Guyatt et al., 1996;
Nakamura et al., 1996),
ClYVV
(Takahashi et al., 1997),
LMV
(Revers et al., 1997)
and SPFMV
(Sakai et al., 1997).
The genome sizes of species of
the genus Potyvirus sequenced to date range from 9496 (TEV) to 10,326
(PRSV) nucleotides excluding the poly(A) tail
(see Shukla et al., 1994).
The length of the 5' non-coding region in different species of this genus ranges
from 85 to 205 nucleotides. Alignment of sequences of this region revealed two
conserved blocks which are referred to as box ‘a’ and ‘b’
(Nicolas and Laliberte, 1992).
The 3' non-coding regions are much more heterogeneous in size (161 to
473 nucleotides) and sequence
(Frenkel et al., 1989;
Berger et al., 1997).
They can be predicted to fold into stable secondary structures
(Turpen, 1989).
Infectious RNA preparations have been obtained from some Potyvirus
species such as SCMV-MDB, PVY, TuMV and TEV
(Pring & Langenberg, 1972;
Makkouk & Gumpf, 1974,
1975;
Hill & Benner, 1976);
TEV-RNA
is reported to contain strands with or without poly(A) which are equally infective
(Hari et al., 1979).
Infectious RNA transcripts have been produced from
full-length cDNA clones to several species of the genus Potyvirus, e.g.
TVMV
(Domier et al., 1989;
Nicolas et al., 1996),
PPV
(Riechmann et al., 1990;
Maiss et al., 1992),
ZYMV
(Gal-On et al., 1991),
PSbMV
(Johansen et al., 1992),
TEV
(Dolja et al., 1992),
PStV
(Flasinski et al., 1996),
PVA
(Puurand et al., 1996),
PVY
(Fakhfakh et al., 1996;
Jakab et al., 1997)
and ClYVV
(Takahashi et al., 1997).
The polyprotein precursor of Potyvirus species is cleaved
by proteinases into 10 different polypeptides
(Dougherty & Carrington, 1988;
Shukla et al., 1991;
Riechmann et al., 1992),
these products
being (Fig. 1): first protein (P1-Pro), helper component (HC-Pro),
third protein
(P3), cylindrical inclusion protein (CI), small nuclear inclusion protein (NIa) which
includes VPg at its N-terminus and the major proteinase (Pro) at its C-terminus,
large nuclear inclusion protein (NIb) and coat protein (CP). VPg and CP are the
only gene products present in virus particles, but P1-Pro, HC-Pro, P3, CI, NIa
and NIb have been detected in infected plants
(Dougherty & Carrington, 1988;
Rodriguez-Cerezo & Shaw, 1991).
Two small putative protein
products (6 kDa each) also occur between P3 and CI and between CI and NIa,
as indicated by the presence of potential cleavage site motifs and the analysis of
in vitro cleavage products, but the former has not been detected in
infected tissue
(Riechmann et al., 1992),
although the latter has
(Restrepo-Hartwig & Carrington, 1994).
Macluravirus species probably have a genomic organization similar to that
of Potyvirus species (Fig. 1) as, like potyviruses,
they are also transmitted
by aphids. The genome of Macluravirus species consists of a single
species of RNA of approximately 8 kb
(Badge et al., 1997).
The
sequences of the C-terminal one-third of the polyproteins of two species, MacMV
and NLV, of this genus presently available show that the coat protein coding
region of the viruses is at the 3'-end of their RNA and a motif similar to DAG,
found in the coat protein N-terminus of Potyvirus species, is also found in
macluraviruses. In MacMV this motif is DAE whereas in NLV it is DVG
(Badge et al., 1997).
In the Potyvirus species TVMV, a mutation from DAG
to DAE gave a non-transmissible product whereas a mutation to DVG left only
minimal transmission function
(Atreya et al., 1990,
1995).
The aphid
transmissibility of the sequenced MacMV and NLV isolates has not been
determined experimentally
(Badge et al., 1997).
The genomic RNA of the Ipomovirus species SPMMV consists of 10,818
nucleotides
(Colinet et al., 1998)
and is slightly longer than that of the
longest RNA of the Potyvirus species PRSV (10,326 nucleotides).
However, the 5' and 3' non-coding regions of SPMMV are 139 and 308
nucleotides long, which are similar to those found in Potyvirus species.
The genome organization and predicted polyprotein cleavage sites of SPPMV
(Colinet et al., 1998)
are very similar to those of Potyvirus species
(see Fig. 1).
The genomic RNA of the Tritimovirus species BrSMV consists of 9672
nucleotides and of WSMV consists of 9384 nucleotides, excluding the poly(A)
tail
(Gotz & Maiss, 1995;
Stenger et al., 1998).
The 5' and 3' non-
coding regions are 145 and 245 nucleotides and 130 and 149 nucleotides long
for BrSMV and WSMV, respectively. Thus, the genome size and length of the 5'
and 3' non-coding regions of Tritimovirus species are very similar to those
of the Potyvirus species. Also, the genome organization and the predicted
cleavage sites of the polyprotein of tritimoviruses are very similar to those of
Potyvirus species
(Gotz & Maiss, 1995;
Stenger et al., 1998).
Specific motifs, described for Potyvirus species polyproteins, are also
almost all present in the polyprotein of tritimoviruses (see Fig. 1).
The genomic RNA of Rymovirus species is expected to have
similar organization to that of the Potyvirus species. Partial
sequence data shows the NIb and CP coding regions are at the 3' end
of the genome.
However, the aphid transmissibility motif found in the N-terminal part of coat
protein of almost all Potyvirus species is not found in the coat protein of
Tritimovirus, Rymovirus or Ipomovirus species. This is not surprising as
species of these latter genera are transmitted by mites or whiteflies.
The complete nucleotide sequences of two Bymovirus species, namely BaYMV (Kashiwazaki et al., 1990, 1991; Davidson et al., 1991; Peerenboom et al., 1992) and BaMMV (Timpe & Kuehne, 1994; Jacobi et al., 1995; Dessens & Meyer, 1996; Kashiwazaki, 1996; Meyer & Dessens, 1996; Peerenboom et al., 1996) are known. The genomes of these viruses consist of two RNA molecules, RNA-1 and RNA-2 (Fig. 1). The RNA-1 (7632-7648 bases in BaYMV and 7263 bases in BaMMV) in both viruses corresponds to the 3' two-thirds of the Potyvirus genome, and codes for proteins analogous to P3, 6K1, CI, 6K2, VPg/NIa, NIb and CP (Kashiwazaki et al., 1990; Peerenboom et al., 1992; Kashiwazaki, 1996; Meyer & Dessens, 1996). The RNA-2 (3585 bases in BaYMV and 3516 bases in BaMMV) codes for a polyprotein which is processed into two mature proteins which have been designated as P1 and P2 (Dessens et al., 1995; Dessens & Meyer, 1996; Kashiwazaki, 1996). The first (P1) shares extensive sequence similarities with HC-Pro of potyviruses (Kashiwazaki et al., 1991; Davidson et al., 1991) and should be called HC-ProLP (helper component-proteinase like protein) to avoid confusion with the unrelated P1 protein of genera with a monopartite genome. The second (P2) is related to the capsid readthrough protein of furoviruses and is believed to play a role in virus transmission by Polymyxa graminis (Dessens et al., 1995; Jacobi et al., 1995; Dessens and Meyer, 1996). The total length of the two BaYMV RNA molecules (11,217 nucleotides) is greater than that of the single Potyvirus genomic RNA due to the presence of 5' and 3' non- coding regions on both RNA molecules (Kashiwazaki et al., 1990, 1991). The 5' non-coding regions of RNA-1 and RNA-2 are very similar in size (171 and 154 nucleotides, respectively) and in sequence (87% identity in the first 154 nucleotides). By contrast, the 3' non-coding regions of RNA-1 and RNA-2 differ in size (231 and 761 nucleotides long, respectively) and show no significant sequence identity (Kashiwazaki et al., 1991). Infectious RNA transcripts have been produced from full-length cDNA clones to BaMMV (Meyer & Dessens, 1997)
Replication
Members of the Potyviridae probably replicate in the cytoplasm. The
replication cycle involves infection, uncoating, translation, polyprotein processing
to yield functional viral-coded products, genome replication via a negative
strand RNA template, progeny virus particle assembly, cell-to-cell movement and
vector transmission from infected to healthy plants. The functions of the gene
products are incompletely known; some gene products, however, have multiple
functions.
The functions of some gene products have been determined by direct evidence,
while those of others have been predicted from sequence similarities with
non-structural proteins of other plant and animal viruses
(Domier et al., 1987;
Dougherty and Carrington, 1988;
Shukla et al., 1991;
Riechmann et al., 1992).
There is little information on the early events in infection. With other plant viruses
it appears that there is usually no virus-host cell receptor recognition system and
that the initial infection process results from virus already present in the seed or
introduced into the potential host by physical damage (insect or fungal vectors or
experimentally by mechanical inoculation). Viral proteins play little, if any, role in
cell entry and viral uncoating is not host-specific
(Matthews, 1991).
The genomic RNA of members of the Potyviridae apparently has no
associated polymerase molecules. Thus, following uncoating, the viral RNA is
translated to generate sufficient virus-coded proteins for subsequent replication,
assembly and spread of progeny virus particles. Although little is known about
the translation process, cap-independent internal initiation of translation has
been proposed for several species of the genus Potyvirus
(Carrington & Freed, 1990;
Nicolaisen et al., 1992;
Levis & Astier-Manifecier, 1993;
Basso et al., 1994).
However, the genomic RNA of PPV
seems to have a leaky scanning mechanism for translation
(Riechmann et al., 1991;
Simon-Buela et al., 1997).
RNA of Potyvirus species is translated as a single polyprotein that is
subsequently cleaved by three distinct proteinases to yield functional viral
products. The C-terminal half
(Dougherty & Carrington, 1988)
of the small
nuclear inclusion protein (NIa) is the major proteinase of potyviruses and is
involved in processing the P3/CI, CI/6kDa, CI/NIa, NIa/NIb and NIb/CP junctions
in the C-terminal two-thirds of the polyprotein
(Carrington & Dougherty 1987;
Carrington et al., 1988;
Hellmann et al., 1988;
Dougherty et al., 1989;
Garcia et al., 1989;
Ghabrial et al., 1990).
The
two 6 kDa products between P3/CI and CI/NIa are also bounded by the NIa
cleavage motifs
(Riechmann et al., 1992).
These primary cleavages
usually occur at susceptible QS, QG or QA bonds
(Shukla et al., 1991),
although occasionally at QV
(Dinant et al., 1991)
or QE bonds
(Johansen et al., 1991;
Gough & Shukla, 1993).
In some cases the Q residue
in the above dipeptide sequences is replaced by E
(Shukla et al., 1994).
Thus, there appears to be a simple cleavage motif, V-x-x-Q(E)-(A,S,G, E or V),
that is common to all members of the Potyviridae
(Shukla et al., 1994).
The NIa proteinase is a cysteine-type proteinase that is probably
structurally related to trypsin-like serine proteinases
(Bazan & Fletterick, 1988).
Its active site is Cys2188 in the TEV polyprotein
(Dougherty & Carrington, 1988).
The processing of the N-terminal third of the Potyvirus polyprotein (P1-
Pro/HC-Pro/P3) does not involve the QG, QA or QS type recognition
sequences that are cleaved by the NIa proteinase in the C-terminal two-thirds of
the precursor. The HC-Pro/P3 cleavage site is G763-G764 in TEV
(Carrington et al., 1989);
equivalent sites exist in other genomic sequences
(Riechmann et al., 1992;
Shukla et al., 1994).
The cleavage is
effected by a second potyvirus-encoded proteinase similar to papain-like
cysteine proteinases
(Kamphuis et al., 1985)
which is in a 20 kDa domain
of the carboxyl-terminal half of the aphid transmission HC-Pro protein
(Carrington et al., 1989).
The active site cysteine and histidine residues
are residues 649 and 722 in the TEV polyprotein
(Oh & Carrington, 1989).
The third proteinase involved in polyprotein processing is the P1-Pro protein, a
serine protease with the active site serine at position 256 in TEV
(Verchot et al., 1991),
which cleaves the P1-Pro/HC-Pro junction at a conserved YS
(304-305 in TEV) or FS bond
(Mavankal & Rhoads, 1991;
Carrington & Herndon, 1992).
The subcellular site(s) of potyviral RNA synthesis has yet to be identified but, as
found with other positive-strand RNA viruses
(Verchot et al., 1991),
is
probably in the cytoplasm. Potyviruses produce no sub-genomic RNA molecules
and the entire RNA genome is copied. Replication thus requires a polymerase, a
primer and an unwinding protein to separate the dsRNA products and enable
multiple copies of progeny strands to be transcribed. The viral proteins involved
are CI, VPg(NIa) and NIb, which form a multicomponent, membrane-associated,
replication complex, similar to that found in picorna-, como- and nepoviruses in
which proteins are coded for by a similarly ordered gene set
(Domier et al., 1987;
Goldbach & Wellink, 1988).
CI is an RNA helicase that, in the
presence of NTP, can unwind RNA duplexes with 3'-overhangs in the 3' to 5'
direction
(Lain et al., 1990,
1991;
Fernandez et al., 1995,
1997).
The VPg
(Shahabuddin et al., 1988)
is the N-terminal half of NIa
(Murphy et al., 1990)
and is attached to the 5' end of the RNA via a phosphate
ester linkage to Y60 (Y1860 in the TVMV polyprotein) in the conserved sequence
motif MNY
(Murphy et al., 1991).
NIb is the RNA-dependent RNA
polymerase of members of the Potyviridae
(Domier et al., 1987;
Martin et al., 1995;
Hong & Hunt, 1996);
it contains the consensus
sequence motif (GDD) of viral RNA-dependent RNA polymerases and is the
most conserved of the Potyviridae gene products.
Nevertheless, it is still unclear
(Riechmann et al., 1992)
(i) how the
parental RNA establishes and organizes the specific replication site within the
cytoplasm; (ii) how the polymerase primes parental RNA to transcribe the
negative sense RNA copies that are used as templates for subsequent viral RNA
synthesis; (iii) what the precise nature of the potyvirus replicative intermediate is;
or (iv) what the significance of the nuclear targeting signals and nuclear
accumulation of VPg(NIa), NIb and the NIa-NIb complex are. NIa and NIb
aggregate as equimolar nuclear inclusions in many, but not all, Potyvirus
infections and both contain nuclear targeting signals
(Restrepo et al., 1990;
Carrington et al., 1991).
The low sequence identity between the P1-Pro proteins of biologically distinct
potyviruses suggest that P1-Pro, particularly its N-terminal, non-proteinase
domain, may be involved in some specific virus-host interaction. The weak
sequence identity between the cell movement protein (P30) of tobacco mosaic
tobamovirus and P1-Pro of some potyviruses suggests that the latter may control
cell-to-cell movement
(Domier et al., 1987),
although the precise role of
P1-Pro has yet to be resolved.
Proteins encoded by the genome of members of the Potyviridae appear to have multiple functions with different functions associated with different protein domains. For example, the central domain of coat protein is required for virus assembly (Jagadish et al., 1991) and cell-to-cell transport (Dolja et al., 1994), the surface-exposed N-terminal domain is involved in aphid transmission (Atreya et al., 1990, 1991) whereas the N- and C- terminal domains mediate long-distance transport (Dolja et al., 1994, 1995). Furthermore, cis-active RNA elements within the CP cistron have been associated with genome amplification (Mahajan et al., 1996; Haldeman-Cahill et al., 1998). Similarly, the N-terminal domain of HC-Pro is involved in aphid-mediated transmission (Atreya et al., 1992), the central region is necessary for efficient genome amplification (Klein et al., 1994; Cronin et al., 1995; Kasschau et al., 1997) and long- distance movement (Cronin et al., 1995; Kasschau et al., 1997) and the C-terminal domain mediates polyprotein processing (Carrington et al., 1989) and possibly cell-to-cell movement (Rojas et al., 1997). The N-terminal domain of NIa acts as VPg (Murphy et al., 1990) and interacts either directly or indirectly with host components to facilitate long-distance movement (Schaad et al., 1997) whereas its C-terminal domain is the major proteinase of members of the Potyviridae (Carrington & Dougherty, 1987). Besides being an RNA helicase (Lain et al., 1990, 1991), the CI protein is also involved in cell-to-cell movement (Andrews & Shalla, 1974; Forster et al., 1988; Carrington et al., 1998) as CI are associated with plasmodesmata (Lawson et al., 1971; Murant & Roberts 1971; Andrews & Shalla, 1974; Rodriquez-Cerezo et al., 1997); moreover, the CI protein is a major component of the replication complex, and ribonucleoprotein complexes probably also facilitate cell transport.
Relationships
Serology
Serological relationships among members of the
Potyviridae are complex. Most definitive potyviruses are serologically
related to at least one and, in many instances, to several other potyviruses.
Serological relationships between many pairs of potyviruses have not been
detected, and other relationships may be only indirect. Thus, unexpected paired
relationships were found between LMV and BYMV, and between BYMV and
BCMV, but not between BCMV and LMV
(Alba & Oliveira, 1976).
Also,
strains of one potyvirus may differ considerably in their serological affinities.
Although a few potyviruses react adequately in agar-gel immunodiffusion tests,
the particles of many must first be fragmented, e.g. by sonication
(Tomlinson et al., 1965),
or the coat protein chemically dissociated into
subunits (e.g. with SDS and/or pyrrolidine)
(Shepard et al., 1974).
However, dissociated proteins are very poor immunogens, and may not react
with antisera to intact particles
(Moghal & Francki, 1976),
and/or fail to
detect the relationships observed between the dissociated particle proteins of
related members of the Potyviridae.
Biochemical and immunochemical investigations of coat proteins have
established the molecular basis for serology in the family Potyviridae and
provided explanations for the occurrence of variable cross-reactivity of polyclonal
antisera, unexpected paired relationships between distinct viruses, and lack of
cross-reactions between some strains
(Shukla et al., 1992b).
Thus it is now known that the N- and C- terminal regions of the coat protein are
on the particle surface (Fig. 2) and can be removed without affecting particle
assembly
(Allison et al., 1985;
Shukla et al., 1988); that the
exposed N termini of the coat proteins are highly variable in length and sequence
(Shukla & Ward, 1988,
1989a,
1989b),
are immunodominant, and
that antibodies to this region are usually virus-specific
(Dougherty et al., 1985;
Shukla et al., 1988,
1989b);
that most cross-reactions
between distinct viruses are attributable to the presence of differing proportions
of antibodies against the conserved core region of the coat protein
(Shukla et al., 1989b;
1989d);
that these cross-reacting antibodies are
increased if degraded virus particles or prolonged injection protocols are
employed; such antibodies are valuable family- or genus-specific probes capable
of detecting species of the genera Potyvirus, Tritimovirus and
Ipomovirus
(Shukla et al., 1989a:
Jordan & Hammond, 1991;
Mishra et al., 1997).
The location, size and sequence of the virus-
specific and group-specific epitopes have been identified by immunochemical
analysis of overlapping synthetic octatapeptides
(Shukla et al., 1989d).
The CI proteins induced by members of the Potyviridae are distinct gene
products which have an RNA helicase function
(Lain et al., 1990,
1991)
and are antigenically unrelated to the coat protein; the CI of unrelated viruses are
also serologically unrelated even when the viruses are propagated in the same
plant species; however, CI proteins of serologically closely related Potyvirus
strains appear to be immunochemically indistinguishable
(Hiebert et al., 1971;
Purcifull et al., 1973).
Cross-protection
Cross-protection occurs between some
Potyvirus species (e.g. TEV, PVY and HMV;
Bawden & Kassanis, 1941)
but
not between different strains of the same Potyvirus species, such as PVY
(Horvath, 1969)
or MDMV
(Shepherd, 1965;
Paulson & Sill, 1970;
Gillaspie & Koike, 1973;
Tosic, 1981).
Two or more potyviruses can occur, and have
synergistic effects, in naturally infected plants
(Hollings & Brunt, 1981b).
Conflicting reports on failed cross-protection may sometimes be
attributed to the incorrect identification of the viruses and strains used. For
example, the SCMV/MDMV group is now know to consist of four distinct viruses
(SCMV, MDMV, JGMV and SrMV)
(Shukla et al., 1989c,
1992a);
similarly, viruses previously referred to as BCMV
(McKern et al., 1992;
Vetten et al., 1992)
and SMV
(Jain et al., 1992) have
also been shown to consist of more than one distinct virus (also see
Shukla et al., 1994).
Coat protein and genome sequences
Several attempts have
been made to differentiate individual potyviruses, to define virus strains, and to
separate potyviruses into sub-groups by using host range, cross-protection,
particle length and serology
(Bos, 1970:
Lindsten et al., 1976),
host range
and serology
(Jones & Diachun, 1977),
serology and amino acid analyses
of the particle protein
(Moghal & Francki, 1976),
and morphology of CI
(Edwardson, 1974).
Sub-groupings suggested by using these criteria show
various anomalies and inconsistencies, and none has been generally accepted
(Francki et al., 1985).
It has been shown that (i) coat protein and gene sequence data are the definitive
criteria for establishing relationships between members of the Potyviridae
(Ward et al., 1992);
(ii) the N-terminal region of coat protein is highly
variable whereas the C-terminal two-thirds of the protein (core region and C-
terminus) is conserved and shows significant sequence identity (>65%) with
other distinct potyviruses
(Shukla & Ward, 1988,
1989a,
1989b;
Ward & Shukla, 1991);
(iii) variation in the coat protein core region is similar to
that of other parts of the genome of Potyvirus species and is a good index
of genetic relatedness
(Shukla et al., 1991);
(iv) coat protein sequence
data can be used to discriminate among members of the Potyviridae and
strains, the bimodal distribution of sequence identities being inconsistent with the
‘continuum’ hypothesis
(Bos, 1970;
Hollings & Brunt, 1981a,
1981b)
which
previously attempted to explain the inability to develop a satisfactory taxonomy of
the family Potyviridae
(Shukla & Ward, 1988;
Ward & Shukla, 1991);
(v) coat protein sequence data can be used to construct a phylogenetic
tree in which the potyvirus ‘group’ is considered to be a family
(Ward & Shukla, 1991;
Barnett, 1991)
divided into five genera, in which distinct viruses
correspond to species and isolates with the highest levels of sequence identity
(>90%) to strains
(Ward & Shukla, 1991);
(vi) strains of some
Potyvirus species, such as those of SCMV and BCMV are now
recognized as distinct viruses
(Shukla et al., 1989c,
1992a;
McKern et al., 1992;
Vetten et al., 1992);
(vii) some members of the genus
Potyvirus previously believed to be distinct species are strains of the
same virus, e.g. PStV, PMMV, BlCMV, AzMV, three soybean isolates from
Taiwan, some strains of CABMV and the serogroup B strains of BCMV are
strains of BCMV
(McKern et al., 1992);
PMV and WLMV are strains of
BYMV
(McKern et al., 1993);
VNV is a strain of WMV 2
(Wang et al., 1993);
and WMV 2 may appear to be a strain of SMV
(Frenkel et al., 1989;
Jain et al., 1992);
(viii) the coat protein sequence data can
also reveal previously unrecognized sub-sets of distinct viruses that are closely
related, for example, ZYMV, SbMV, WMV 2, PWV, BCMV, BCMNV (formerly the
serotype A strains of BCMV) form one sub-set
(Ward & Shukla, 1991;
Ward et al., 1992);
BYMV and ClYVV form another; (ix) the 3' non-coding region
of the potyviral genome is a good marker of genetic relatedness
(Frenkel et al., 1989,
1992);
(x) although any part of the viral genome can be used to
establish phylogenetic relationships, the coat protein or 3' non-coding regions are
the most convenient
(Shukla et al., 1991;
Ward & Shukla, 1991).
The coat protein and genome sequences have provided an hierarchical
classification of members of the Potyviridae. Initially the family
Potyviridae was divided into four genera, Potyvirus, Rymovirus,
Bymovirus and tentatively Ipomovirus
(Barnett, 1991)
on the basis of
sequence diversity
(Ward & Shukla, 1991),
and by coincidence these four
genera corresponded to the four modes of vector transmission, aphid, mite,
fungus and whitefly, respectively. At the time the only sequence available for the
mite-transmitted viruses was that of WSMV, but since RGMV had been
described first, the name Rymovirus was selected for this genus.
Recent sequence data
(Niblett et al., 1991;
Gotz & Maiss, 1995;
Schubert & Rabenstein, 1995;
Salm et al., 1996a)
has revealed that the
mite-transmitted potyviruses fall into two groups and a new genus Tritimovirus
was approved to accommodate WSMV, BrSMV and the possible species
SCSMV. The remaining species (AgMV, HoMV, ONMV, RGMV and SpMV)
have been retained as a distinct sixth genus (Rymovirus) by the
ICTV Potyviridae Study Group. As shown in
Fig. 3, sequence analyses divide the
Potyviridae into only five groupings not six, with the rymoviruses
sharing strong sequence identity with members of the Potyvirus
genus. Thus sequence identities no longer correlate with the current
ICTV classification of the Potyviridae into six genera. It is our
view that this classification is in error and that AgMV, RGMV, ONMV,
HoMV and SpMV should be transferred from the Rymovirus genus
to the Potyvirus genus and the Rymovirus genus be
removed and made redundant from the family Potyviridae.
Such a proposal is currently being discussed by the Potyviridae Study
Group.
Affinities with Other Groups
Members of the Potyviridae are readily differentiated from other filamentous viruses of the genera Allexivirus, Capillovirus, Carlavirus, Closterovirus, Crinivirus, Foveavirus, Potexvirus, Trichovirus and Vitivirus. Their particles are more flexuous than those of allexiviruses, carlaviruses, foveaviruses and potexviruses but less so than those of capilloviruses, closteroviruses, criniviruses, trichoviruses and vitiviruses. Their genome organization indicates that they belong to the picorna-like supergroup of viruses whose RNAs have a VPg covalently bound to the 5' end, a poly(A) tail at the 3' end, and are expressed as a single polyprotein which is subsequently cleaved by proteinases to yield several functional proteins, including a conserved ordered gene set of non-structural proteins that are involved in RNA replication (Goldbach & Wellink, 1988).
Figures

Genome organization of the Potyvirus (TEV) and Bymovirus (BaYMV) genera of the Potyviridae family. AI, amorphous inclusion; CI, cylindrical inclusion; NIa and NIb, small and large nuclear inclusion proteins which aggregate in the nucleus to form a nuclear inclusion body; CP, coat protein. P1-Pro and HC-Pro cleave the bonds between P1 and HC-Pro (helper component proteinase) and HC-Pro and P3 proteins, respectively while the NIa- Pro cleaves the rest of the polyprotein bonds (filled diamonds); VPg, genome- linked protein covalently attached to the 5' terminal nucleotide (represented by filled circle); HC-ProLP, HC-Pro-like protein. Modified from Berger et al. (1998).

Schematic drawing showing the linear sequence of the coat protein subunit, the subunit folding pattern, the surface location of the N- and C-termini and the assembly of PVY particle (based on Shukla & Ward, 1989a).
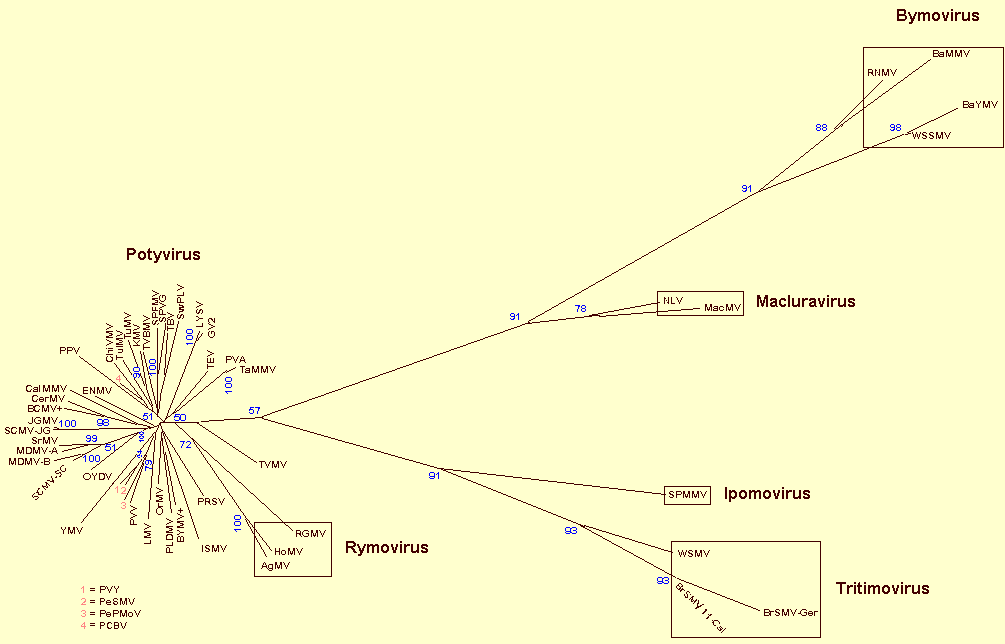
Phylogenetic analysis of coat protein amino acid sequences of members of the family Potyviridae using the FM method (modified from Berger et al., 1997). The family is divided currently into six genera, namely Potyvirus, Macluravirus, Ipomovirus, Tritimovirus, Rymovirus and Bymovirus. As shown here the mite-transmitted Rymovirus species, AgMV, HoMV and RGMV (enclosed in a box), cluster with the aphid-transmitted species of the genus Potyvirus. On the basis of this and other information we believe their inclusion in a separate genus Rymovirus is no longer justified. The Potyviridae Study Group are looking at this proposal to remove the Rymovirus genus and place these mite transmitted viruses in the genus Potyvirus. Other mite-transmitted viruses now form the new Tritimovirus genus. The ‘BCMV +’ represents the bean common mosaic virus subgroup consisting of BCMV (of which AzMV, BlCMV, DeMV, PStV are strains), BCMNV, CABMV (includes SAPV), WMV-2 (includes VNV), PWV and ZYMV, and the ‘BYMV +’ refers to the bean yellow mosaic virus subgroup consisting of BYMV (includes pea mosaic virus) and ClYVV.
References list for DPV: Potyviridae family (366)
- Alba & Oliveira, Summa Phytopath. 2: 178, 1976.
- Aleman, Marcos, Brugidou, Beachy & Fauquet, Arch. Virol. 141: 1259, 1996.
- Allison, Dougherty, Parks, Willis, Johnston, Kelly & Armstrong, Virology 147: 309, 1985.
- Allison, Johnston & Dougherty, Virology 154: 9, 1986.
- Ammar, Jarlfors & Pirone, Phytopathology 84: 1054, 1994.
- Andrews & Shalla, Phytopathology 64: 1234, 1974.
- Atkinson & Slykhuis, Can. Pl. Dis. Sur. 43: 154, 1963.
- Atreya, Raccah & Pirone, Virology 178: 161, 1990.
- Atreya, Raccah & Pirone, Proc. natl Acad. Sci. USA 88: 7887, 1991.
- Atreya, Atreya, Thornbury & Pirone, Virology 191: 106, 1992.
- Atreya, Lopez-Moya, Chu, Atreya & Pirone, J. gen. Virol. 76: 265, 1995.
- Badge, Robinson, Brunt & Foster, J. gen. Virol. 78: 253, 1997.
- Barnett, Arch. Virol. 118: 139, 1991.
- Basso, Dallaire, Devantier & Laliberte, J. gen. Virol. 75: 3157, 1994.
- Baunoch, Das & Hari, J. gen. Virol. 71: 2479, 1990.
- Bawden & Kassanis, Ann. appl. Biol. 28: 107, 1941.
- Bazan & Fletterick, Proc. natl Acad. Sci. USA 85: 7872, 1988.
- Berger & Pirone, Virology 153: 256, 1986.
- Berger, Wyatt, Shiel, Silbernagel, Druffel & Mink, Arch. Virol. 142: 1979, 1997.
- Berger, Barnett, Brunt, Colinet, Edwardson, Hammond, Hill, Jordan, Kashiwazaki, Makkouk, Morales, Rybicki, Spence, Ohki, Uyeda, van Zaayen & Vetten, 7th Rep. ICTV, 1998 (in press).
- Blanc, Lopez-Moya, Wang, Garcia-Lampasona, Thornbury & Pirone, Virology 231: 141, 1997.
- Bond & Pirone, Phytopathology 60: 437, 1970.
- Bos, Neth. J. Pl. Path. 76: 8, 1970.
- Brakke, Skopp & Lane, Phytopathology 80: 1401, 1990.
- Brunt, CMI/AAB Descr. Pl. Viruses 170, 4 pp., 1976.
- Brunt, Ann. appl. Biol. 87: 355, 1977.
- Brunt & Atkey, Ann. appl. Biol. 78: 339, 1974.
- Carrington & Dougherty, J. Virol. 61: 2540, 1987.
- Carrington & Freed, J. Virol. 64: 1590, 1990.
- Carrington & Herndon, Virology 187: 302, 1992.
- Carrington, Cary & Dougherty, J. Virol. 62: 2313, 1988.
- Carrington, Freed & Sanders, J. Virol. 63: 4459, 1989.
- Carrington, Freed & Leinicke, Pl. Cell 3: 953, 1991.
- Carrington, Jensen & Schaad, Pl. J. 14: 393, 1998.
- Chamberlain & Catherall, Ann. appl. Biol. 85: 105, 1977.
- Choi & Wakimoto, Ann. Phytopath. Soc. Japan 45: 32, 1979.
- Christie & Edwardson, Monograph Ser. Fla agric. Exp. Stn 9: 150 pp., 1977.
- Chu, Johnson, Thornbury, Black & Pirone, Virus Genes 10: 283, 1995.
- Colinet, Kummert & Lepoivre, Virus Res. 53: 187, 1998.
- Cronin, Verchot, Haldeman-Cahill, Schadd & Carrington, Pl. Cell 7: 549, 1995.
- Damirdagh & Shepherd, Phytopathology 60: 132, 1970.
- Davidson, Prols, Schell & Steinbiss, J. gen. Virol. 72: 989, 1991.
- De Mejia, Hiebert & Purcifull, Virology 142: 24, 1985a.
- De Mejia, Hiebert & Purcifull, Thornbury & Pirone, Virology 142: 34, 1985b.
- Dinant, Lot, Albouy, Kuziak, Meyer & Astier-Manifacier, Arch. Virol. 116: 235, 1991.
- Dessens & Meyer, Virus Genes 12: 95, 1996.
- Dessens, Nguyen & Meyer, Arch. Virol. 140: 325, 1995.
- Dolja, McBride & Carrington, Proc. natl Acad. Sci. USA 89: 10208, 1992.
- Dolja, Haldeman, Robertson, Dougherty & Carrington, EMBO J. 13: 1482, 1994.
- Dolja, Haldeman-Cahill, Montgomery, VandenBosch & Carrington, Virology 207: 1007, 1995.
- Domier, Franklin, Shahabuddin, Hellmann, Overmeyer, Hiremath, Siaw, Lomonossoff, Shaw & Rhoads, Nucleic Acids Res. 14: 4517, 1986.
- Domier, Shaw & Rhoads, Virology 158: 20, 1987.
- Domier, Franklin, Hunt, Rhoads & Shaw, Proc. natl Acad. Sci. USA 86: 3509, 1989.
- Dougherty & Carrington, Ann. Rev. Phytopath. 26: 123, 1988.
- Dougherty, Wills & Johnston, Virology 144: 66, 1985.
- Dougherty, Cary & Parks, Virology 171: 356, 1989.
- Edwardson, Am. J. Bot. 53: 359, 1966.
- Edwardson, Monograph Ser. Fla agric. Exp. Stn 4: 398 pp., 1974.
- Edwardson, Christie & Ko, Phytopathology 74: 1111, 1984.
- Fakhfakh, Vilaine, Makni & Robaglia, J. gen. Virol. 77: 519, 1996.
- Fang, Allison, Zambolim, Maxwell & Gilbertson, Virus Res. 39: 13, 1995.
- Fernandez, Lain & Garcia, Nucleic Acids Res. 23: 1327, 1995.
- Fernandez, Guo, Saenz, Simon-Buela, de Cedron & Garcia, Nucleic Acids Res. 25: 4474, 1997.
- Flasinski, Gunasinghe, Gonzales & Cassidy, Gene 171: 299, 1996.
- Forster, Bevan, Harbison & Gardner, Nucleic Acids Res. 16: 291, 1988.
- Francki, Milne & Hatta, Atlas of Plant Viruses, CRC Press, Vol. 2, 183, 1985.
- Francki, Fauquet, Knudson & Brown, Arch. Virol, [Suppl. 2], pp. 351-356, 1991.
- Frenkel, Ward & Shukla, J. gen. Virol. 70: 2775, 1989.
- Frenkel, Jilka, Shukla & Ward,. J. Virol. Methods 36: 51, 1992.
- Gal-On, Antignus, Rosner & Raccah, J. gen. Virol. 72: 2693, 1991.
- Gal-On, Antignus, Rosner & Raccah, J. gen. Virol. 73: 2183, 1992.
- Garcia, Riechmann & Lain, Virology 170: 362, 1989.
- Ghabrial, Smith, Parks & Dougherty, J. gen. Virol. 71: 1921, 1990.
- Gillaspie & Koike, Phytopathology 63: 1300, 1973.
- Goldbach & Wellink, Intervirology 29: 260, 1988.
- Goodman, McDonald, Horne & Bancroft, Phil. Trans. R. Soc., Ser. B. 276: 173, 1976.
- Gotz & Maiss, J. gen. Virol. 76: 2053, 1995.
- Gough & Shukla, Intervirology 136: 181, 1993.
- Govier & Kassanis, Virology 61: 420, 1974.
- Govier & Woods, J. gen. Virol. 13: 127, 1971.
- Govier, Kassanis & Pirone, Virology 78: 306, 1977.
- Gunasinghe, Flasinski, Nelson & Cassidy, J. gen. Virol. 75: 2519, 1994.
- Guyatt, Proll, Menssen & Davidson, Arch. Virol. 141: 1231, 1996.
- Haldeman-Cahill, Daros & Carrington, J. Virol. 72: 4072, 1998.
- Hari, Siegel, Rozek & Timperlake, Virology 92: 568, 1979.
- Hellmann, Shaw & Rhoads, Virology 163: 554, 1988.
- Hiebert & McDonald, Virology 56: 349, 1973.
- Hiebert, Purcifull, Christie & Christie, Virology 43: 638, 1971.
- Hill & Shepherd, Virology 47: 806, 1972.
- Hill & Benner, Virology 75: 419, 1976.
- Hinostroza-Orihuela, Virology 67: 276, 1975.
- Hollings & Brunt, in Handbook of Plant Virus Infections and Comparative Diagnosis, ed. E. Kurstak, Amsterdam: Elsevier/North Holland, p. 731, 1981a.
- Hollings & Brunt, CMI/AAB Descriptions of Plant Viruses 245, 7 pp., 1981b.
- Hollings, Stone & Bock, CMI/AAB Descriptions of Plant Viruses 162, 4 pp., 1976.
- Hong & Hunt, Virology 226: 146, 1996.
- Horvath, Zentbl. Bakt. ParasitKde Abt. 2. 123: 249, 1969.
- Hull, Milne & van Regenmortel, Arch. Virol. 120: 151, 1991.
- Husted, Bech, Albrechtsen & Borkhardt, Phytopathology 84: 161, 1994.
- Huth, Lesemann & Paul, Phytopath. Z. 111: 37, 1984.
- Huttinga & Mosch, Neth. J. Pl. Path. 80: 19, 1974.
- Inouye & Fujii, CMI/AAB Descriptions of Plant Viruses 172, 4 pp., 1977.
- Inouye & Saito, CMI/AAB Descriptions of Plant Viruses 143, 4 pp., 1975.
- Jacobi, Peerenboom, Schenk, Antoniw, Steinbiss & Adams, Virus Res. 37: 99, 1995.
- Jagadish, Ward, Gough, Tulloch, Whittaker & Shukla, J. gen. Virol. 72: 1543, 1991.
- Jain, McKern, Tolin, Hill, Barnett, Tosic, Ford, Beachy, Yu, Ward & Shukla, Phytopathology 82: 294, 1992.
- Jakab, Droz, Brigneti, Baulcombe & Malnoe, J. gen. Virol. 78: 3141, 1997.
- Jayaram, Hill & Miller, J. gen. Virol. 73: 2076, 1992.
- Johansen, Rasmussen, Heide & Borkhardt, J. gen. Virol. 72: 2652, 1991.
- Johansen, Kohnen, Dougherty & Hampton, Phytopathology 82: 1111, 1992.
- Johansen, Keller, Dougherty & Hampton, J. gen. Virol. 77: 1329, 1996.
- Jones & Diachun, Phytopathology 67: 831, 1977.
- Jordan & Hammond, J. gen. Virol. 72: 25, 1991.
- Kamphuis, Drenth & Baker, J. Molec. Biol. 182: 317, 1985.
- Kashiwazaki, Arch. Virol. 141: 2077, 1996.
- Kashiwazaki, Minobe, Omura & Hibino, J. gen. Virol. 71: 2781, 1990.
- Kashiwazaki, Minobe & Hibino, J. gen. Virol. 72: 995, 1991.
- Kassanis, Ann. appl. Biol. 26: 705, 1939.
- Kasschau, Cronin & Carrington, Virology 228: 251, 1997.
- Khetarpal, Belkacem & Maury, in: Ph.D. Thesis by Khetarpal, Univ. Southern Paris, pp. 49-54, 1989.
- Kitajima & Costa, J. gen. Virol. 20: 413, 1973.
- Kitajima & Lovisolo, J. gen. Virol. 16: 265, 1972.
- Klein, Klein, Rodriguez-Cerezo, Hunt & Shaw, Virology 204: 759, 1994.
- Knuhtsen, Hiebert & Purcifull, Virology 61: 200, 1974.
- Koenig & Plese, CMI/AAB Descriptions of Plant Viruses 239, 3 pp., 1981.
- Kusaba, Toyama, Yumoto & Takobe, Bull. Tottori Agric. Exp. Stn. 2: 208 pp., 1971.
- Lain, Riechmann, Martin & Garcia, Virus Res. 10: 325, 1989.
- Lain, Riechmann & Garcia, Nucleic Acids Res. 18: 7003, 1990.
- Lain, Martin, Riechmann & Garcia, J. Virol. 63: 1, 1991.
- Langenberg & Schroeder, Virology 55: 218, 1973.
- Langenberg & Schroeder, J. gen. Virol. 23: 51, 1974.
- Lawson & Hearon, Virology 44: 454, 1971.
- Lawson, Hearon & Smith, Virology 46: 453, 1971.
- Layne, Meth. Enzym. 3: 447, 1957.
- Levis & Astier-Manifacier, Virus Genes 7: 367, 1993.
- Liao, Chien, Chung, Ching & Han, J. agric. Res. China 28: 127, 1979.
- Lindsten, Brishammar & Tomenius, Meddn. St. VaxtskAnst. 16: 289, 1976.
- Mahajan, Dolja & Carrington, J. Virol. 70: 4370, 1996.
- Maiss, Trimpe, Brisske, Jelkmann, Casper, Himmler, Mattanovich & Katinger, J. gen. Virol. 70: 513, 1989.
- Maiss, Timpe, Brisske-Rose, Lesemann & Casper, J. gen. Virol. 73: 709, 1992.
- Makkouk & Gumpf, Phytopathology 64: 1115, 1974.
- Makkouk & Gumpf, Virology 63: 336, 1975.
- Martin, Cervera & Garcia, Virus Res. 37: 127, 1995.
- Matthews, Intervirology 12: 258, 1979.
- Matthews, Plant Virology, 3rd Edition, pp. 338-378, Academic Press, New York, 1991.
- Mavankal & Rhoads, Virology 185: 721, 1991.
- McDonald & Bancroft, J. gen. Virol. 35: 251, 1977.
- McDonald & Hiebert, Virology 58: 200, 1974.
- McDonald, Beveridge & Bancroft, Virology 69: 327, 1976.
- McKern, Mink, Barnett, Mishra, Whittaker, Silbernagel, Ward & Shukla, Phytopathology 82: 923, 1992.
- McKern, Barnett, Whittaker, Mishra, Strike, Xiao, Ward & Shukla, Phytopathology 83: 355, 1993.
- Mernaugh, Gardner & Yacom, Virology 106: 273, 1980.
- Meyer & Dessens, Virology 219: 268, 1996.
- Meyer & Dessens, J. gen. Virol. 78: 3147, 1997.
- Mishra, McKimm-Breschkin, Xiao & Shukla, Ind. J. Virol. 13: 15, 1997.
- Moghal & Francki, Virology 73: 350, 1976.
- Mumford & Seal, J. Virol. Methods 69: 73, 1997.
- Murant & Roberts, J. gen. Virol. 10: 65, 1971.
- Murphy, Rhoads, Hunt & Shaw, Virology 178: 285, 1990.
- Murphy, Rychlik, Rhoads, Hunt & Shaw, J. Virol. 65: 511, 1991.
- Nakamura, Honkura, Iwai, Ugaki & Ohashi, Ann. Phytopath. Soc. Japan 62: 472, 1996.
- Niblett, Zagula, Calvert, Kendall, Stark, Smith, Beachy & Lommel, J. gen. Virol. 72: 499, 1991.
- Nicolaisen, Johansen & Poulsen, FEBS Lett. 303: 169, 1992.
- Nicolas & Laliberte, J. gen. Virol. 73: 2785, 1992.
- Nicolas, Pirone & Hellmann, Arch. Virol. 141: 1535, 1996.
- Oh & Carrington, Virology 173: 692, 1989.
- Oshima, Tanaka & Sako, Arch. Virol. 141: 1991, 1996.
- Paliwal, Arch. Virol. 63: 123, 1980.
- Palkovics, Burgyan & Balazs, Virus Genes 7: 339, 1993.
- Paulsen & Sill, Pl. Dis. Reptr 54: 627, 1970.
- Peerenboom, Prols, Schell, Steinbiss & Davidson, J. gen. Virol. 73: 1303, 1992.
- Peerenboom, Jacobi, Antoniw, Schlichter, Cartwright, Steinbiss & Adams, Virus Res. 40: 149, 1996.
- Pirone & Blanc, Annu. Rev. Phytopath. 34: 227, 1996.
- Pirone & Thornbury, Phytopathology 73: 872, 1983.
- Plese & Stefanac, Mitt. biol. BundAnst. Ld- u. Forstw. 170: 47, 1976.
- Plumb & Jones, J. gen. Virol. 18: 407, 1973.
- Pring & Langenberg, Phytopathology 62: 253, 1972.
- Purcifull, Hiebert & McDonald, Virology 55: 275, 1973.
- Puurand, Makinen, Paulin & Saarma, J. gen. Virol. 75: 457, 1994.
- Puurand, Valkonen, Makinen, Rabenstein & Saarma, Virus Res. 40: 135, 1996.
- Restrepo-Hartwig & Carrington, J. Virol. 68: 2388, 1994.
- Restrepo, Freed & Carrington, Pl. Cell 3: 987, 1990.
- Revers, Yang, Walter, Soche, Lot, Le Conde, Candresse & Dunez, Virus Res. 47: 167, 1997.
- Riechmann, Lain & Garcia, Virology 177: 710, 1990.
- Riechmann, Lain & Garcia, Virology 185: 544, 1991.
- Riechmann, Lain & Garcia, J. gen. Virol. 73: 1, 1992.
- Robaglia, Durand-Tardif, Tronchet, Boudazin, Astier-Manifacier & Casse-Delbart, J. gen. Virol. 70: 935, 1989.
- Rodriguez-Cerezo & Shaw, Virology 185: 572, 1991.
- Rodriguez-Cerezo, Findlay, Shaw, Lomonossoff & Qiu, Virology 236: 296, 1997.
- Rojas, Zerbini, Allison, Gilbertson & Lucas, Virology 237: 283, 1997.
- Rubio-Huertos & Lopez-Abella, Microbiol. esp. 19: 77, 1966.
- Saito, Ueda & Inaba, Ann. Phytopath. Soc. Japan 32: 87, 1966.
- Saito, Tsuchiazaki & Hibino, Ann. Phytopath. Soc. Japan 34: 347, 1968.
- Sakai, Mori, Morishita, Tanaka, Hanada, Usugi & Nishiguchi, Arch. Virol. 142: 1553, 1997.
- Salm, Rey & French, Arch. Virol. 141: 185, 1996a.
- Salm, Rey & Rybicki, Arch. Virol. 141: 2237, 1996b.
- Schaad, Lellis & Carrington, J. Virol. 71: 8624, 1997.
- Schubert & Rabenstein, Eur. J. Pl. Path. 101: 123, 1995.
- Shahabuddin, Shaw & Rhoads, Virology 163: 635, 1988.
- Shepard & Carroll, J. Ultrastruct. Res. 21: 145, 1967.
- Shepard, Secor & Purcifull, Virology 58: 464, 1974.
- Shepherd, Phytopathology 55: 1250, 1965.
- Shukla & Ward, J. gen. Virol. 69: 2703, 1988.
- Shukla & Ward, Adv. Virus Res. 36: 273, 1989a.
- Shukla & Ward, Arch. Virol. 106: 171, 1989b.
- Shukla, Strike, Tracy, Gough & Ward, J. gen. Virol. 69: 1497, 1988.
- Shukla, Ford, Tosic, Jilka & Ward, Arch. Virol. 105: 143, 1989a.
- Shukla, Jilka, Tosic & Ford, J. gen. Virol. 70: 13-23, 1989b.
- Shukla, Tosic, Jilka, Ford, Toler & Langham, Phytopathology 79: 223, 1989c.
- Shukla, Tribbick, Mason, Hewish, Geysen & Ward, Proc. natl Acad. Sci. USA 86: 8192, 1989d.
- Shukla, Frenkel & Ward, Can. J. Pl. Path. 13: 178, 1991.
- Shukla, Frenkel, McKern, Ward, Jilka, Tosic & Ford, in Potyvirus Taxonomy, ed. O.W. Barnett, Arch. Virol. [Suppl. 5]: 363, 1992a.
- Shukla, Lauricella & Ward, in Potyvirus Taxonomy, ed. O. W. Barnett, Arch. Virol. [Suppl. 5]: 57, 1992b.
- Shukla, Ward and Brunt, The Potyviridae. CAB International, UK, 1994.
- Simon-Buela, Guo & Garcia, Virology 233: 157, 1997.
- Singh & Singh, Can. J. Pl. Path. 18: 209, 1996.
- Slykhuis, Phytopathology 60: 319, 1970.
- Slykhuis, CMI/AAB Descriptions of Plant Viruses 18, 4 pp., 1973.
- Slykhuis, Andrews & Pittman, Can. J. Pl. Sci. 37: 113, 1957.
- Stenger, Hall, Choi & French, Phytopathology 88: (in press) 1998.
- Takahashi, Takahashi & Uyeda, Virus Genes 14: 235, 1997.
- Teycheney, Tavert, Delbos, Ravelonandro & Dunez, Nucleic Acids Res. 23: 10115, 1989.
- Thole, Dalmay, Burgyan & Balazs, Gene 123: 149, 1993.
- Thornbury, Patterson, Dessens & Pirone, Virology 178: 573, 1990.
- Timpe & Kuehne, Eur. J. Pl. Path. 100: 233, 1994.
- Toler & Hebert, Phytopathology 54: 428, 1964.
- Tomlinson, Walkey, Hughes & Watson, Nature, Lond. 207: 495, 1965.
- Tosic, Agronomie 1: 83, 1981.
- Tsuneyoshi, Matsumi, Natsuaki & Sumi, Arch. Virol. 143: 97, 1998.
- Turpen, J. gen. Virol. 70: 1951, 1989.
- Usugi, Kashiwazaki, Omura & Tsuchizaki, Ann. Phytopath. Soc. Japan 55: 26, 1989.
- Vance, Moore, Turpen, Bracker & Hollowell, Virology 191: 106, 1992.
- Varma, Gibbs, Woods & Finch, J. gen. Virol. 2: 107, 1968.
- Veerisetty, Intervirology 11: 167, 1979.
- Verchot, Koonin & Carrington, Virology 185: 527, 1991.
- Vetten, Lesemann & Maiss, in Potyvirus Taxonomy, ed. O. W. Barnett, Arch. Virol. [Suppl. 5]: 415, 1992.
- Von Wechmar, Chauhan & Knox, in Potyvirus Taxonomy, ed. O. W. Barnett, Arch. Virol. [Supp. 5]: 415, 1992.
- Wang, Ammar, Thornbury, Lopez-Moya & Pirone, J. gen. Virol. 77: 861, 1996.
- Wang, Beck, Gardner & Pearson, Arch. Virol. 129: 93, 1993.
- Ward & Shukla, Intervirology 32: 269, 1991.
- Ward, McKern, Frenkel & Shukla, in Potyvirus Taxonomy, ed. O. W. Barnett, Arch. Virol. [Supp. 5]: 382, 1992.
- Weintraub, Agrawal & Ragetli, Can. J. Bot. 61: 855, 1973.
- Weintraub, Ragetli & Lo, J. Ultrastruct. Res. 46: 131, 1974.
- Wisler, Purcifull & Hiebert, J. gen. Virol. 76: 37, 1995.
- Yeh, Jan, Chiang, Doong, Chen, Chung & Bau, J. gen. Virol. 73: 2531, 1992.
- Zitter & Tsai, Pl. Dis. Reptr 61: 1024, 1977.
- Namba, Kashiwazaki, Lu, Tamura & Tsuchizaki, Arch. Virol. 143: 631, 1998.