Details of DPV and References
DPV NO: 391 April 2002
Family: Unallocated ssRNA+ viruses
Genus: Benyvirus
Species: Beet necrotic yellow vein virus | Acronym: BNYVV
This is a revised version of DPV 144
Beet necrotic yellow vein virus
T. Tamada Research Institute for Bioresources, Okayama University, Kurashiki, Okayama, 710-0046 JAPAN
Contents
- Introduction
- Main Diseases
- Geographical Distribution
- Host Range and Symptomatology
- Strains
- Transmission by Vectors
- Transmission through Seed
- Transmission by Grafting
- Transmission by Dodder
- Serology
- Nucleic Acid Hybridization
- Relationships
- Stability in Sap
- Purification
- Properties of Particles
- Particle Structure
- Particle Composition
- Properties of Infective Nucleic Acid
- Molecular Structure
- Genome Properties
- Satellite
- Relations with Cells and Tissues
- Ecology and Control
- Notes
- Acknowledgements
- Figures
- References
Introduction
First described by Canova (1966) in Italy, and by Tamada & Baba (1973) in Japan.
- Selected synonyms
- Beet rhizomania virus
A virus with straight tubular particles with a diameter of about 20 nm and five modal lengths in the range 80 to 390 nm, containing five ssRNA species which are referred to as RNAs 1 to 5. RNAs 1 and 2 encode "house-keeping" genes involved in replication, assembly and cell-to-cell movement, whereas RNAs 3, 4 and 5 are associated with vector-mediated infection and disease development in sugar beet roots. RNA 5 is found only in certain isolates that occur in limited areas. The virus has a narrow host range and is transmitted by the protozoan Polymyxa betae and by mechanical inoculation. It occurs wherever sugar beets are grown and causes a disease of economic importance.
Main Diseases
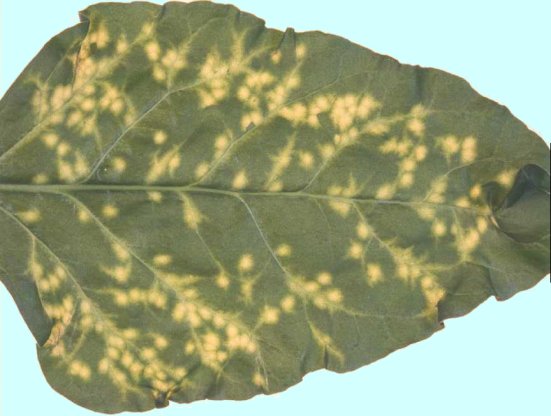
The virus causes "rhizomania" disease of sugar beet (Beta vulgaris var. saccarifera). In nature, it infects sugar beet, fodder beet, Swiss chard and spinach. The disease is usually distributed as foci (patches) in the sugar beet field (Fig. 1). The most useful leaf symptom is visible at the end of the growing season. Leaves become pale green in colour, with long petioles and upright growth (Fig. 2). Symptoms are characterized by root stunting and proliferation of lateral rootlets on the main tap roots (Fig. 3), and yellow-brown coloration of vascular bundles (Fig. 4). These typical root symptoms give rise to the name "rhizomania". In early and severe infection, the plants are stunted, wilted and eventually die. In this condition, the bright yellow colour (Fig. 5) followed by necrosis along veins (giving the virus name "necrotic yellow vein") is very rarely seen. This symptom only results from movement of virus to leaves. Infection late in the season may produce no obvious symptoms.
The virus causes severe damage in sugar beets. Yield losses depend greatly on the inoculum level in the soil, the weather conditions and the time of infection. Severe infection leads to a reduction in root yield of 50 % or more, and the sugar content is greatly decreased. Unusually low levels of sugar are used as an early indicator of virus infection (Asher, 1993).
Geographical Distribution
The virus is probably distributed worldwide wherever there are major sugar beet industries. It was first found in Italy during the 1950s, in the Po plain and Adige valley (Canova, 1959). From 1971 to 1990, it was observed in an increasing number of countries, from central and southern Europe to eastern and northern Europe and Middle East Asia (Austria, Belgium, Bulgaria, Croatia, Czech Republic, France, Germany, Greece, Hungary, Iran, Kazakstan, Kirgizia, Netherlands, Poland, Roumania, Russian Federation, Slovakia, Spain, Switzerland, Turkey, Ukraine, United Kingdom, Yugoslavia) (Asher, 1993; Tamada, 1999). It was found in Sweden in 1997 (Lennefors et al., 2000). In Japan, the virus was first found in 1965 (Kanzawa & Ui, 1972). In China, since the first recorded finding in Nei Menggu in 1978 (Gao et al., 1983), it has spread throughout the districts along the Yellow River, Xinjiang Urgur and Heilongjiang. In the USA, the virus was first recorded in California in 1983 (Duffus et al., 1984) and in Texas in 1987 (Duffus & Liu, 1987). Between 1992 and 1994, it was found in Colorado, Idaho, Nebraska, Wyoming and Minnesota (Rush & Heidel, 1995).
Host Range and Symptomatology
The virus has a narrow host range. It is transmitted by inoculation of sap to most species of the family Chenopodiaceae and several species belonging to the Aizoaceae, Amaranthaceae, Caryophyllaceae and Solanaceae (Tamada & Baba, 1973; Kuszala & Putz, 1977; Horvath, 1994; Hugo et al., 1996; T. Tamada, unpublished data). Many of these plants are hosts of the vector Polymyxa betae (Abe & Ui, 1986; Abe & Tamada, 1986; Barr & Asher, 1992, 1996; Hugo et al., 1996). The virus tends to be restricted to the inoculated leaves of host plants; it becomes systemic in Beta macrocarpa, some accessions of B. vulgaris spp. maritima, spinach and Nicotiana benthamiana (Tamada, 1975; T. Tamada, unpublished data).
Diagnostic speciesBeta vulgaris (sugar beet). Inoculated leaves develop chlorotic lesions 6-8 days after inoculation. The lesions then become bright yellow (Fig. 6), enlarge and tend to coalesce, spreading along the veins. Few plants are systemically infected; they show chlorotic or yellow spotting, yellow vein-banding, vein necrosis, leaf distortion, wilting and stunting (Fig. 7).
Beta macrocarpa. Inoculated leaves develop yellowish local lesions, followed by systemic yellow mottle or yellow flecks with severe stunting (Fig. 8).
Chenopodium quinoa and C. amaranticolor. Chlorotic or necrotic lesions appear in inoculated leaves 5-7 days after inoculation. Not systemically infected.
Tetragonia expansa. Inoculated leaves usually develop bright yellow lesions (Fig. 9). However, chlorotic, yellowish, or necrotic lesions appear sometimes in inoculated leaves, depending on the RNA species contained in the virus isolate inoculated (Fig. 9). This species can be used for distinguishing virus isolates of different pathogenicity (see Strains).
Tetragonia expansa and Chenopodium quinoa (inoculated leaves) can be used for maintaining cultures and as sources of virus.
Tetragonia expansa, Chenopodium quinoa and C. amaranticolor are good local lesion hosts.
Sugar beet (Beta vulgaris) seedlings are useful for testing transmission by the vector.
Strains
Although no serological differences are found among virus isolates, the majority have been classified into A and B types based on sequence differences (Kruse et al.,1994; Koenig et al., 1995; Miyanishi et al., 1999) (see Relationships). Virus isolates containing RNA 5 are found in Japan, China, France and Kazakstan (Koenig et al., 1997b; Miyanishi et al., 1999; Koenig & Lennefors, 2000). In Japan, about half of the isolates tested contained RNA 5. Virus isolates containing RNA 5 are more virulent than isolates lacking RNA 5 (Tamada et al., 1996b).
Different strains of the virus are characterized on the basis of the type of local lesions in Tetragonia expansa (Fig. 9) (Tamada, 1975; Tamada et al., 1989): YS (bright yellow spots), CS (chlorotic spots), fCS (faint chlorotic spots) and NS (necrotic spots). This depends on the presence of RNA 3, RNA 4 and/or RNA 5, or their deletion mutants. RNA3-containing isolates produce YS type lesions and cause "rhizomania" symptoms in sugar beet roots (see Main Diseases), whereas isolates without RNA3 produce CS type lesions and do not cause "rhizomania" symptoms (Tamada et al., 1999). Isolates that contain RNA5 but lack RNA3 cause severe CS type lesions (Tamada et al., 1989). The A and B types are not distinguished from each other by lesion type in T. expansa, but RNA5-containing isolates, including the P type strains (see Relationships), produce diffuse local lesions, sometimes with necrosis (T. Tamada, unpublished data). Deletions in each of the small RNAs occur after multiplication in mechanically inoculated leaves of T. expansa or C. quinoa (Kuszala et al., 1986; Koenig et al., 1986; Tamada et al., 1989; Bouzoubaa et al., 1991). Such deletion mutants also affect symptom phenotypes, and pathogenicity to sugar beet (Jupin et al., 1991, 1992; Tamada et al., 1999).
Transmission by Vectors
Transmitted by the plasmodiophorid protozoan Polymyxa betae. When sugar beet plants are grown in infested fields, zoospores of the protozoa are released from cystosori or resting spores (Fig. 10) and infect root hairs or epidermal cells of the roots (Keskin, 1964; Keskin & Fuchs, 1969; Barr & Asher, 1996). At that time, rootlets become infected with the virus carried by viruliferous zoospores. After penetration of the protozoa into the root cells, plasmodia develop into zoosporangia, from which new zoospores are released within a few days (Keskin, 1964; Dirven & Peters, 1995). Plasmodia of P. betae may also form cystosori, which can survive in the soil for many years. The virus replicates in plant cells, but there is no evidence for multiplication in P. betae (Abe & Tamada, 1986). After multiplication, the virus is acquired by the protozoa, presumably through the membrane of the thallus, and may accumulate in the plasmodia. Virus-like particles are observed within numerous vacuoles in young immature plasmodia or zoospores (Fig. 11) (Tamada, 1975; Abe & Tamada, 1986; Rysanek et al., 1992).
Transmission through Seed
Not transmitted.
Serology
Moderately immunogenic in rabbits; antisera with titres of 1/1024 in ring precipitin tests have been obtained. Several rat or mouse monoclonal antibodies to purified virus particles have been obtained (Torrance et al., 1988; Koenig et al., 1990). The coat protein has been analyzed in detail by immunosorbent electron microscopy (ISEM) (Lesemann et al., 1990), following trypsin treatment (Koenig et al., 1990), and by using synthetic overlapping peptides (Commandeur et al., 1994). Such tests indicate that virus particles have five different epitopes (Commandeur et al., 1992). ISEM and ELISA are useful for virus detection (Koenig et al., 1984; Putz et al., 1988). Immunogold-silver labelling (Giunchedi & Poggi-Pollini, 1988; Poggi-Pollini & Giunchedi, 1989; Scholten et al., 1994) or tissue print immunoassay (Kaufmann et al., 1992) are used for examining localization and distribution of virus in root tissues of sugar beet.
Nucleic Acid Hybridization
Nucleic acid hybridization assays have been developed for the detection of the virus (Richards et al., 1985; Koenig et al., 1986; Putz et al., 1988; Saito et al., 1997). RT-PCR methods also are useful for virus detection (Henry et al., 1995; Fenby et al., 1995), and for distinguishing virus strains or variants which contain different smaller RNAs and their deletion mutants (T. Tamada, unpublished data).
Relationships
Serological differences have not been detected among isolates of the virus (Kuszala et al., 1986; Torrance et al., 1988). However, the majority of isolates may be classified into two groups (A and B types) on the basis of single-strand conformation polymorphisms of immunocapture RT-PCR products (Kruse et al., 1994; Koenig et al., 1995). The A type is found in most European countries, the USA, China and Japan, whereas the B type is detected in Germany, France (Kruse et al., 1994), Japan (Miyanishi et al., 1999) and Sweden (Lennefors et al., 2000). The nucleotide sequences of French (B type) and Japanese (A type) isolates are 97 % identical when averaged over all four RNAs (Saito et al., 1996). The two types can be discriminated by three common amino acid changes in the coat protein gene (Miyanishi et al., 1999). Three groups of RNA 5 variants are found in virus isolates from Japan, China and France, most of which belong to the A type (Miyanishi et al., 1999). French RNA 5-containing isolates, which were named P type (Koenig et al., 1995, 1997b), are very closely related to the A type (Miyanishi et al., 1999). P type isolates have also been detected in Kazakstan (Koenig & Lennefors, 2000). There are some differences among A, B and P types in pathogenicity and virus multiplication in different sugar beet cultivars (Heijbroek et al., 1999).
Beet necrotic yellow vein, Beet soil-borne mosaic, Burdock mottle and Rice stripe necrosis viruses are placed in the genus Benyvirus based on genome organization and sequence homology (Heidel et al., 1997; Tamada, 1999; Hirano et al., 1999; Morales et al., 1999; Lee et al., 2001). Although Rice stripe necrosis virus is listed as a tentative species of the genus Furovirus in the 7th ICTV report, this virus (Colombian isolate) contains four RNA species, in which the two larger RNAs are similar in size to the corresponding RNAs of BNYVV and Beet soil-borne mosaic virus (Morales et al., 1999). In addition, the 3' ends of their RNAs are polyadenylated (Morales et al., 1999). BNYVV and Beet soil-borne mosaic virus are serologically related but distinct (Wisler et al., 1994). Cross-protection between the two viruses has been found in sugar beet (Mahmood & Rush, 1999). BNYVV and Burdock mottle virus are not serologically related (Tamada, 1999). The coat proteins of BNYVV and Beet soil-borne mosaic virus are 56 % identical (Lee et al., 2001), whereas those of BNYVV and Burdock mottle virus are 38 % identical (Hirano et al., 1999). Thus, BNYVV and Beet soil-borne mosaic virus are more closely related to each other than to Burdock mottle virus.
Stability in Sap
In sugar beet sap, the thermal inactivation point (10 min) is 65 to 70°C and dilution end-point about 10 -4 (Tamada & Baba, 1973; Putz, 1977). Infectivity is retained for 5 days at 20°C and for 8 days at 4°C. Infectivity of sap extracts is decreased greatly by freezing (Tamada & Baba, 1973).
Purification
The virus particles can be purified with a modification of the methods described by Koenig et al. (1984) and Tamada et al. (1989). Use inoculated leaves of Tetragonia expansa or Chenopodium quinoa with well developed lesions. Grind fresh or frozen leaves in 3 to 5 volumes of 0.5 M borate buffer (pH 9.0), containing 0.001 M Na2EDTA, and squeeze through cheesecloth. After low speed centrifugation for 10 min, add Triton X-100 to 2% to the supernatant and stir for 5 min. Purify the virus particles by centrifugation through a 20% sucrose cushion and resuspend the pellets in 0.05 M borate (or 0.05 M Tris-HCl) buffer (pH 8.0). Purify further by centifugation through a sucrose density gradient in the same buffer. Collect the light-scattering virus zone (usually a broad band) and concentrate by high speed centrifugation. Resuspend the final pellets in 0.01 M borate buffer (pH8.0), 0.01 M Tris-HCl buffer (pH 8.0) or water. For antiserum production, purify the virus further by caesium chloride density gradient centrifugation.
Particle Structure
The virus particles are rod-shaped, with helical symmetry and a central canal. Their diameter is about 20 nm and the modal lengths are 390, 265, 105, 90 and 80 nm, corresponding to five RNAs (RNAs 1 to 5, respectively) (Fig. 12) (Tamada, 1975; Putz, 1977; Tamada et al., 1989). The single-start right-handed helix has a 2.6 nm pitch with an axial repeat of 4 turns, involving 49 subunits of a 21K protein (Steven et al., 1981).
Particle Composition
Nucleic acids: The viral RNA consists of five distinct species with lengths of 6746 (RNA 1), 4612 (RNA 2), 1773-1774 (RNA 3), 1465-1467 (RNA 4) and 1342-1347 (RNA 5) nucleotides (Fig. 13) (Bouzoubaa et al., 1985, 1986, 1987; Saito et al., 1996; Kiguchi et al., 1996). All five viral RNAs have cap structures at their 5'-termini and terminate in 3'-poly(A) tails. Close sequence homology among the five RNAs is limited to the 5'-terminal 8-9 nucleotides and the last 70 nucleotides preceding the 3'-poly(A) tail. RNAs 3, 4 and 5 have unusually long 5'-noncoding regions of 445, 379 and 443 nucleotides, respectively.
Protein: A single coat protein species consisting of 188 amino acids, Mol. Wt 21,000 (Putz, 1977).
Genome Properties
RNAs 1 and 2 encode "house-keeping" genes involved in replication, particle assembly and cell-to-cell movement, whereas RNAs 3, 4 and 5 are associated with vector-mediated infection and disease development in sugar beet roots (Fig. 13) (Lemaire et al., 1988; Richards & Tamada, 1992).
Accession numbers for nucleotide sequences of genome parts of BNYVV isolates
CP: coat protein, TGB: triple gene block
RNA 1 contains a single long open reading frame (ORF), potentially encoding a polypeptide of 237K, which contains the information necessary for replication of the viral genome (Fig. 13). This ORF contains three distinct replication-associated domains: a methyltransferase domain, an NTP-binding/helicase domain and a polymerase domain. The primary in vitro translation product of RNA 1 is a 220K polypeptide, which can be cleaved autocatalytically into two species of 150K and 66K; a papain-like protease motif occurs between the helicase domain and the polymerase domain (Hehn et al., 1997).
RNA 2 contains six ORFs (Fig. 13). The 5'-proximal ORF encodes the 21K coat protein. The coat protein cistron is separated from a long (54K) in-phase ORF by a single amber termination codon which is suppressed about 10 % of the time to produce a coat protein-54K fusion protein of 75K (readthrough protein) (Ziegler et al., 1985). This readthrough protein is a minor component of virions and is located at the ends of the viral particles (Haeberle et al., 1994). A region within the N-terminal half of the readthrough domain is involved in virus particle assembly (Schmitt et al., 1992). The C-terminal portion contains sequences important for vector transmission (Tamada & Kusume, 1991; Tamada et al., 1996a), including a KTER motif identified by alanine scanning mutagenesis (Tamada et al., 1996a). The central portion of RNA 2 contains a cluster of three slightly overlapping genes known as the triple gene block (TGB), encoding proteins of 42K, 13K and 15K. Synthesis of the 42K protein is directed by subgenomic RNA 2 suba, whereas synthesis of both 13K and 15K proteins is probably directed by a dicistronic subgenomic mRNA, RNA 2 subb (Gilmer et al., 1992a). The TGB proteins have amino acid sequence homologies and similar hydrophobicities to equivalent proteins involved in cell-to-cell movement of a number of other plant viruses, including the potex-, carla-, hordei- pomo- and pecluviruses. The first TGB protein (42K) has sequence motifs characteristic of a superfamily 1 DNA or RNA helicase, including a "P-Loop" ATP/GTP-binding domain (Koonin & Dolja, 1993). This protein possesses nucleic acid-binding activity (Bleykasten et al., 1996). The binding site is situated near the N-terminus, but the P-Loop motif is not required for nucleic acid-binding. The second TGB protein (13K) has two potentially membrane-spanning hydrophobic domains separated by a hydrophilic sequence that contains a highly conserved peptide motif of unknown significance. The N-terminal portion of the third TGB protein (15K) is hydrophobic. The 42K and 13K proteins are detected in a membrane-enriched subcellular fraction (Niesbach-Klosgen et al., 1990). Expression of the first two proteins can be complemented independently, but over- expression of the 15K protein inhibits cell-to-cell movement of the virus (Bleykasten-Grosshans et al., 1997). Highly specific interactions are found among the TGB proteins, which are important for their function and/or stability (Lauber et al., 1998a). The 3'-proximal ORF of RNA 2 encodes a cysteine-rich 14K protein which is expressed from another subgenomic RNA (Gilmer et al., 1992a). The 14K protein is soluble (Niesbach-Klosgen et al., 1990) and may bind RNA and/or DNA to regulate host genome expression; there is evidence that the 14K protein acts in cis to stimulate the accumulation of RNA 2, but also acts independently and in trans as an enhancer of viral coat protein synthesis (Hehn et al., 1995).
RNA 3 encodes a 25K protein (Fig. 13), which is soluble in vivo (Niesbach-Klosgen et al., 1990) and localized in both cytoplasm and nuclei of cells in infected leaves (Haeberle & Stussi-Garaud, 1995). It increases virus multiplication in roots and is responsible for "rhizomania" symptoms (Koenig et al., 1991; Tamada et al., 1999). RNA 3 also has dramatic effects on symptoms in leaves (Tamada, 1975; Kuszala et al, 1986; Jupin et al., 1992); virus isolates containing RNA 3 produce bright yellow local lesions in Tetragonia expansa or Chenopodium quinoa, whereas isolates lacking RNA 3 produce much milder symptoms (Fig. 9). Mutagenic analysis revealed that the 25K protein is responsible for this yellow spot lesion phenotype (Jupin et al., 1992). The 3'-terminal 600 nucleotides of RNA-3 are easily detected in vivo as an unencapsidated, subgenomic mRNA, which encodes a 4.6K polypeptide of unknown function (Bouzoubaa et al., 1991). In addition, there is a short ORF, called N (necrosis), which overlaps the C-terminus of the 25K ORF (Fig. 13) (Jupin et al., 1992). The N gene is not detectably expressed from full-length RNA 3, but can induce necrotic local lesions when activated by deletion of upstream sequences, or when inserted into another plant virus replicon. RNA 3 is essential for systemic (vascular) movement in Beta macrocarpa (Tamada et al., 1989), but this may depend on an RNA 3 sequence domain rather than an RNA 3-coded protein (Lauber et al., 1998b).
RNA 4 has an ORF for a 31K protein (Fig. 13), which is soluble in vivo (Niesbach-Klosgen et al., 1990). RNA 4 is important for fungus transmission (Tamada & Abe, 1989). RNA 5 contains a single ORF encoding a 26K protein, which is involved in the severity of symptom expression in roots (Fig. 13) (Tamada et al., 1996b). Synergistic interactions are found between RNA 3 and RNA 4 or RNA 5 and RNA 4 (Tamada et al., 1989; Richards &Tamada, 1992; T. Tamada, unpublished data).
The smaller RNAs are useful for the identification of sequences that are recognized by the viral replication process. At the 3'-terminus, the cis-active domain is located within the last 70 residues preceding the 3'-poly(A) tail, which region can be folded into a double hairpin secondary structure common to the five viral RNAs (Jupin et al., 1990). This structure probably comprises the promoter for initiation of minus-strand RNA synthesis. At the 5'-terminus, cis-active sequence elements are located in the first 292 residues of RNA 3, in which at least two of the essential subdomains appear to be important in the formation of secondary structure (Gilmer et al., 1992b, 1993). A sequence about 200 nucleotides from the 5'-terminus contains the encapsidation signal of RNA 3 (Gilmer et al., 1992b). For RNA 4, the 5'-proximal cis-essential elements are limited to the first 400 residues (Gilmer et al., 1992b). These sequences are presumably important for initiation of synthesis of the RNA plus- strand.
Relations with Cells and Tissues
Virus particles are encountered in loose, small aggregates and scattered in the cytoplasm of infected cells of leaves or roots (Tamada, 1975; Putz & Vuittenez, 1980; Russo et al., 1981; Giunchedi et al., 1981). Particles are often in angle-layer aggregates (Fig. 14), in which the angles between virus particles are 45° and 90°. There are no large well-ordered aggregates and no characteristic inclusion bodies. No association is observed between virus particles and cell organelles (Putz & Vuittenez, 1980), but in infected cells there is an increase in the number of rough endoplasmic reticulum profiles (Salle et al., 1986). Immunogold-silver labelling (Scholten et al., 1994) showed that, in rootlets of sugar beets that were infected by P. betae, the virus is detected in the epidermis, cortex parenchyma, endodermis and interstitial parenchyma, but usually not in vascular tissues. Kaufmann et al. (1992), however, showed by tissue print immuno-blotting that the virus is occasionally detected in xylem vessels. Virus-like particles are seen in sections of young plasmodia, zoosporangia and immature zoospores of P. betae (Fig. 11), but not in mature zoospores (Tamada, 1975; Abe & Tamada, 1986; Rysanek et al., 1992).
Ecology and Control
The virus is not transmitted by seed or pollen, but its spread can result from soil contamination of seeds which have been produced in infested areas. Dried infected roots or air-dried soils have been shown to retain infectivity for more than 15 years (Abe & Tamada, 1986). Similar longevity is observed in field conditions. Thus, the disease has occurred when sugar beets have been grown in fields in which no crops have been cultivated for 10 to 15 years (Schlosser, 1988). Although P. betae is able to infect several weed plants with the virus (Abe & Tamada, 1986; Barr & Asher, 1992), common arable weeds probably play only a minor role in the epidemiology of the virus (Barr & Asher, 1996; Hugo et al., 1996). The spread of the virus (in resting spores of viruliferous P. betae) is brought about by the use of machinery on contaminated land, by transportation of infested soil (beet roots, potatoes, possibly beet seed or vegetables grown on infested land) and by irrigation (Richard-Molard, 1985; Heijbroek, 1988). Factory waste, washing water and agricultural equipment are potentially significant in the spread of the disease. Stable manure can also play a role in the dispersal of the virus (Heijbroek, 1988). The resting spores of the vector can also be dispersed by flood or wind.
The most important factors affecting the development of disease in infected fields are the level of inoculum, and soil temperature and moisture (Schlosser, 1988; Asher, 1993). P. betae requires a high soil moisture level for maximum activity. The disease becomes severe where there is poor soil structure, inadequate drainage, frequent heavy rainfall or the use of (excessive) irrigation. The relatively high temperature of about 25 °C is optimal for P. betae activity (Blunt et al., 1991); therefore, soil temperature in the spring and early summer may be of particular importance for the disease severity (Blunt et al., 1992). P. betae infects most rapidly and actively in neutral or alkaline soils (Abe, 1987).
Control in infested fields is not likely to be achieved by management of cultural or agronomic practices. Precautionary measures should be taken to avoid contamination of healthy areas and to limit the spread of the disease in areas already infested. In areas where the virus is still absent or the inoculum potential is low, measures such as wider crop rotation, controlled irrigation and effective drainage can delay the spread and incidence of the disease (Asher, 1993). Also, the disease incidence may be reduced by early sowing (Blunt et al., 1992), transplanting in paper pots (Richard-Molard, 1985) and decreasing soil pH (Abe, 1987). Some chemicals, especially soil fumigants, have been partially effective in reducing inoculum levels in the soil, but are generally not economic (Schlosser, 1988; Asher, 1993). In heavily infested fields in the USA, however, the use of fumigation (Telone) and of tolerant cultivars have been effective in reducing the damage (Martine & Whitney, 1990; Harveson & Rush, 1994).
The cultivation of resistant or tolerant cultivars is, therefore, the most promising way to control the disease (Schlosser, 1988; Asher, 1993). Sources of virus resistance have been described, mainly originating from wild beet, Beta vulgaris spp. maritima (Lewellen et al., 1987; Whitney, 1989; Geyl et al., 1995). Since the introduction of the first partially resistant cultivar Rizor (de Biaggi, 1987), several new cultivars have been developed with higher levels of resistance and are presently grown in rhizomania-infested regions. Two resistance genes, which are referred to as Rz1, from B. vulgaris spp. vulgaris accession Holly-1-4 (Pelsy & Merdinoglu, 1996; Scholten et al., 1996; Barzen et al., 1997), and Rz2, from B. vulgaris spp. maritima accession WB42 (Scholten et al., 1996), have been identified by RAPD or SCAR markers. In general, the resistance of cultivars is caused by a restriction of virus multiplication and/or translocation in the roots, but not by resistance to infection with the vector P. betae (Giunchedi et al., 1987; Poggi-Pollini & Giunchedi, 1989; Bürcky & Büttner, 1991; Paul et al., 1992, Scholten et al., 1994; Tamada et al., 1999). The planting of resistant cultivars may be expected to delay the build up of virus inoculum in soil (Tuitert et al., 1994). Two wild beet species, B. patellaris and B. procumbens, are reported to be resistant to P. betae (Barr et al., 1995). As regards pathogen-derived resistance, sugar beet plants transformed with the BNYVV coat protein gene have shown resistance to the virus in the greenhouse and in the field (Mannerlöf et al., 1996).
Notes
The benyviruses (type species Beet necrotic yellow vein virus) were formerly classified as possible members of the Furovirus genus, which have rigid rod-shaped virions, plus-strand RNA genomes, are generally bipartite and are transmitted by soil-borne pathogens of the family Plasmodiophorales. Based on genome organization and sequence relatedness, these viruses have been recently re-classified in the following genera (Torrance & Mayo, 1997): Furovirus (type species Soil-borne wheat mosaic virus), Pecluvirus (type species Peanut clump virus), Pomovirus (type species Potato mop-top virus) and Benyvirus. These genera have not been assigned to a family or families. Benyvirus differs from these and other genera of viruses with rod-shaped particles, including Tobamovirus, Tobravirus and Hordeivirus, in polymerase phylogeny, genome organization and expression strategy. For example, BNYVV RNA is 3'-polyadenylated, whereas the 3'-ends of the other virus RNAs are not polyadenylated but can be folded into a tRNA-like structure. The putative replication-associated protein of BNYVV RNA is encoded by a single long ORF producing a large polypeptide, which is autocatalytically processed to give the two final products (Hehn et al., 1997). In contrast, the other viruses contain motifs for these functions on two ORFs separated by a leaky stop codon or on two different RNAs. Based on polymerase phylogeny, Benyvirus is included in a cluster together with the animal Togavirus genus in the alphavirus-like superfamily (Tamada, 1999).
Beet soil-borne mosaic virus is very similar to BNYVV in transmission by Polymyxa betae, host range, particle morphology and genome organization, but the viruses are distinct serologically (Wisler et al., 1994; Rush et al., 1994; Rush and Heidel, 1995). Beet soil-borne mosaic virus is widely distributed in the USA (Rush and Heidel, 1995), but has not been identified from other countries. In greenhouse tests, Beet soil-borne mosaic virus is less virulent than BNYVV (Heidel et al., 1997). Systemic foliar symptoms caused by Beet soil-borne mosaic virus are slight leaf distortion, mottling and yellow vein-banding, and appear more frequently than those caused by BNYVV. Beet soil-borne mosaic virus and BNYVV may be found in the same field and even in the same plant in "rhizomania"-infested areas.
Beet soil-borne virus is also similar to BNYVV in having a common vector and host range (Henry et al., 1986), but is included in the genus Pomovirus, which differs in genome structure and sequence (Koenig et al., 1996, 1997a). Beet soil-borne virus is very common in sugar beet fields throughout the world (Lindsten, 1989) and may be a part of the "rhizomania" disease complex in some fields. Beet soil-borne virus is restricted to roots of sugar beet (no systemic infection) and usually causes no obvious symptoms.
In the field, foliar and root symptoms of BNYVV in sugar beet plants are easily confused with other causes such as some soil-borne fungal pathogens, nematodes, insect-transmitted viruses, or poor soil conditions. For example, foliage symptoms are easily confused with nitrogen deficiency. The discoloration (orange or reddish brown) of the central stele may be confused with symptoms of Fusarium root rot (Rush and Heidel, 1995). Because foliage and root symptoms are thus obscure, the most efficient and accurate diagnosis is done by ELISA. Samples should be extracted from lateral roots or from the tip of the taproots (Giunchedi et al., 1987; Büttner & Bürcky, 1990). In soil or adherent soil, a biological test is possible. Beet bait plants are grown in suspected soil, and their rootlets are tested by ELISA (Beemster & de Heij, 1987; Büttner & Bürcky, 1990). Such bait plant tests can also be used to estimate the level of infestation in the field (Tuitert, 1990).
BNYVV shows a great capacity for local dispersal from infested fields and for spread to regions previously uninfested (Tuitert & Hofmeester, 1992, 1994). Although the exact origin of the epidemic in the 1970s is obscure, it is certain that large areas of beet cultivation in Europe, the Middle East, Asia and the USA were free from the virus until recently, but there has subsequently been considerable spread within Europe and elsewhere. It is thus only in the last 20 to 30 years that the virus has developed into a serious problem in many sugar beet growing regions of the world (Asher, 1993; Tamada, 1999).
Figures

Yellow patches in a sugar beet field infected with beet necrotic yellow vein virus in Hokkaido in September.

Local lesions in inoculated leaves of Tetragonia expansa: (left) healthy; (centre) S-4 (RNA-1+2+4), chlorotic lesions; (right) S-34 (RNA-1+2+3+4), bright yellow lesions.
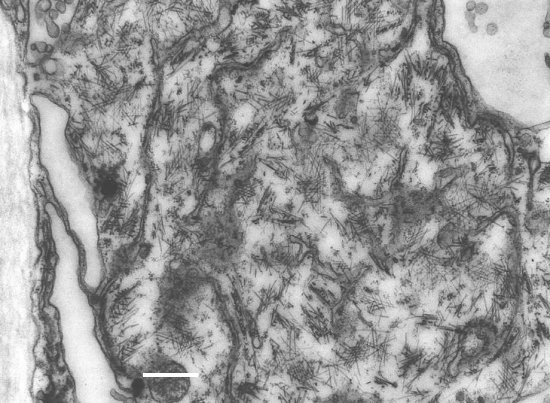
Ultrathin section of immature zoospore of Polymyxa betae from infected roots, showing virus-like particles in the cytoplasm. Bar represents 500 nm.
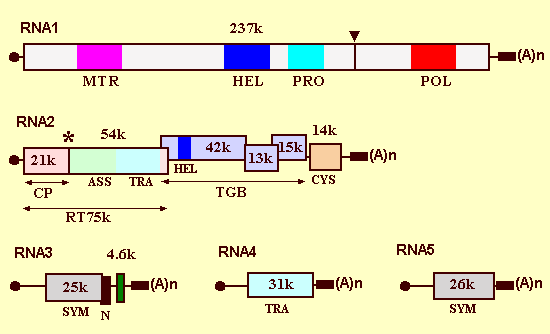
The genome organization of Beet necrotic yellow vein virus. The filled circles indicate the cap structures, the boxes indicate ORFs, and the black rectangles indicate conserved common sequences preceding the 3'-poly(A) tail. The triangle indicates the location of the predicted protease cleavage site and the asterisk indicates a readthrough amber termination codon. The coat protein (CP), the readthrough protein (RT) and the triple gene block (TGB) are shown by arrows. The methyltransferase domain (MTR), the NTP-binding/helicase domains (HEL), the papain-like proteinase domain (PRO), the GDD replicase domain (POL), the cysteine-rich protein (CYS), and regions involved in virus assembly (ASS), transmission (TRA), symptom expression (SYM) and necrosis (N) are indicated.
References list for DPV: Beet necrotic yellow vein virus (391)
- Abe, Report of Hokkaido Prefectural Agricultural Experiment Station, 60, 1987.
- Abe & Tamada, Annals of the Phytopathological Society of Japan 52: 235, 1986.
- Abe & Ui, Annals of the Phytopathological Society of Japan 52: 394, 1986.
- Asher, in The Sugar Beet Crop: Science into practice, p. 311, D. A. Cooke & R. K. Scott, eds, London: Chapman and Hall, 1993.
- Barr & Asher, Plant Pathology 41: 64, 1992.
- Barr & Asher, Mycological Research 100: 203, 1996.
- Barr, Asher & Lewis, Plant Pathology 44: 301, 1995.
- Barzen, Stahl, Fuchs, Borchardt & Salamini, Molecular Breeding 3: 231, 1997.
- Beemster & de Heij, Netherlands Journal of Plant Pathology 93: 91, 1987.
- Bleykasten, Gilmer, Guilley, Richards & Jonard, Journal of General Virology 77: 889, 1996.
- Bleykasten-Grosshans, Guilley, Bouzoubaa, Richards & Jonard, Molecular Plant-Microbe Interactions 10: 240, 1997.
- Blunt, Asher & Gilligan, Plant Pathology 40: 257, 1991.
- Blunt, Asher & Gilligan, Plant Pathology 41: 148, 1992.
- Bouzoubaa, Guilley, Jonard, Richards & Putz, Journal of General Virology 66: 1553, 1985.
- Bouzoubaa, Ziegler, Beck, Guilley, Richards & Jonard, Journal of General Virology 67: 1689, 1986.
- Bouzoubaa, Quillet, Guilley, Jonard & Richards, Journal of General Virology 68: 615, 1987.
- Bouzoubaa, Niesbach-Klosgen, Jupin, Guilley, Richards & Jonard, Journal of General Virology 72: 259, 1991.
- Bürcky & Büttner, Journal of Phytopathology 131: 1, 1991.
- Büttner & Bürcky, Zeitschrift für Pflanzenkrankheiten und Pflanzenschutz 97: 56, 1990.
- Canova, Informatore Fitopatologica 9: 390, 1959.
- Canova, Informatore Fitopatologica 16: 235, 1966.
- Commandeur, Koenig, Lesemann, Torrance, Burgermeister, Liu, Schots, Alric & Grassi, Journal of General Virology 73: 695, 1992.
- Commandeur, Koenig, Manteuffel, Torrance, Luddecke & Frank, Virology 198: 282, 1994.
- De Biaggi, in Proceedings of the 50th Winter Congress of the International Institute for Sugar Beet Research, II, p157, Brussels: IIBR, 1987.
- Dirven & Peters, Journal of Phytopathology 143: 537, 1995.
- Duffus & Liu, Plant Disease 71: 557, 1987.
- Duffus, Whitney, Larsen, Liu & Lewellen, Plant Disease 68: 251, 1984.
- Fenby, Scott, Slater & Elliott, Cellular and Molecular Biology 41: 639, 1995.
- Gao, Deng, Zhai, Ling & Liu, Acta Phytopathologica Sinica 13: 1, 1983.
- Geyl, Garcia Heriz, Valentin, Hehn & Merdinoglu, Plant Pathology 44: 819, 1995.
- Gilmer, Bouzoubaa, Hehn, Guilley, Richards & Jonard, Virology 189: 40, 1992a.
- Gilmer, Richards, Jonard & Guilley, Virology 190: 55, 1992b.
- Gilmer, Allmang, Ehresmann, Guilley, Richards, Jonard & Ehresmann, Nucleic Acids Research 21: 1389, 1993.
- Giunchedi & Poggi-Pollini, Phytopathologica Mediterranea 27: 1, 1988.
- Giunchedi, Langenberg & Marani, Phytopathologica Mediterranea 20: 112, 1981.
- Giunchedi, de Biaggi & Poggi-Pollini, Phytopathologica Mediterranea 26: 23, 1987.
- Haeberle & Stussi-Garaud, Journal of General Virology 76: 643, 1995.
- Haeberle, Stussi-Garaud, Schmitt, Garaud, Richards, Guilley & Jonard, Archives of Virology 134: 195, 1994.
- Harveson & Rush, Plant Disease 78: 1197, 1994.
- Hehn, Bouzoubaa, Bate, Twell, Marbach, Richards, Guilley & Jonard, Virology 210: 73, 1995.
- Hehn, Fritsch, Richards, Guilley & Jonard, Archives of Virology 142: 1051, 1997.
- Heidel, Rush, Kendall, Lommel & French, Plant Disease 81: 1070, 1997.
- Heijbroek, Netherlands Journal of Plant Pathology 94: 9, 1988.
- Heijbroek, Musters & Schoone, European Journal of Plant Pathology 105: 397, 1999.
- Henry, Jones & Coutts, Plant Pathology 35: 585, 1986.
- Henry, Barker, Morris & Hugo, Journal of Virological Methods 54: 15, 1995.
- Hirano, Kondo, Maeda & Tamada, in Proceedings, 4th Symposium of the International Working Group on Plant Viruses with Fungal Vectors, p33, Denver: American Society of Sugar Beet Technologists, 1999.
- Horvath, Acta Phytopathologica et Entomologica Hungarica 29: 109, 1994.
- Hugo, Henry & Harju, Plant Pathology 45: 662, 1996.
- Jupin, Richards, Jonard, Guilley & Pleij, Virology 178: 273, 1990.
- Jupin, Tamada & Richards, Seminars in Virology 2: 121, 1991.
- Jupin, Guilley, Richards & Jonard, The EMBO Journal 11: 479, 1992.
- Kanzawa & Ui, Annals of the Phytopathological Society of Japan 38: 434, 1972.
- Kaufmann, Koenig & Lesemann, Archives of Virology126: 329, 1992.
- Keskin, Archiv fur Mikrobiologie 49: 348, 1964.
- Keskin & Fuchs, Archiv fur Mikrobiologie 68: 218, 1969.
- Kiguchi, Saito & Tamada, Journal of General Virology 77: 575, 1996.
- Koenig & Lennefors, Archives of Virology 145: 1561, 2000.
- Koenig, Lesemann & Burgermeister, Phytopathologishe Zeitschrift 111: 244, 1984.
- Koenig, Burgermeister, Weich, Sebald & Kothe, Journal of General Virology 67: 2043, 1986.
- Koenig, Commandeur, Lesemann, Burgermeister, Torrance, Grassi, Alric, Kallerhoff & Schots, Journal of General Virology 71: 2229, 1990.
- Koenig, Jarausch, Li, Commandeur, Bugermeister, Gehrke & Luddecke, Journal of General Virology 72: 2243, 1991.
- Koenig, Luddecke & Haeberle, Journal of General Virology 76: 2051, 1995.
- Koenig, Beier, Commandeur, Luth, Kaufmann & Luddecke, Virology 216: 202, 1996.
- Koenig, Commandeur, Loss, Beier, Kaufmann & Lesemann, Journal of General Virology 78: 469, 1997a.
- Koenig, Haeberle & Commandeur, Archives of Virology 142: 1499, 1997b.
- Koonin & Dolja, Critical Reviews in Biochemistry and Molecular Biology 28: 375, 1993.
- Kruse, Hoffmann, Koenig, Kaufmann, Commandeur, Solovyev, Savenkov & Burgermeister, Journal of General Virology 75: 1835, 1994.
- Kuszala & Putz, Annales de Phytopathologie 9: 435, 1977.
- Kuszala, Ziegler, Bouzoubaa, Richards, Putz, Guilley & Jonard, Annals of Applied Biology 109: 155, 1986.
- Lauber, Bleykasten-Grosshans, Erhardt, Bouzoubaa, Jonard, Richards & Guilley, Molecular Plant-Microbe Interactions 11: 618, 1998a.
- Lauber, Guilley, Tamada, Richards & Jonard, Journal of General Virology 79: 385, 1998b.
- Lee, Telford, Barren, Scholthof & Rush, Archives of Virology 146: 2443, 2001.
- Lemaire, Merdinoglu, Valentin, Putz, Ziegler-Graff, Guilley, Jonard & Richards, Virology 162: 232, 1988.
- Lennefors, Lindsten & Koenig, Europian Journal of Plant Pathology 106: 199, 2000.
- Lesemann, Koenig, Torrance, Buxton, Boonekamp, Peters & Schots, Journal of General Virology 71: 731, 1990.
- Lewellen, Skoyen & Erichsen, in Proceedings of the 50th Winter Congress of the International Institute for Sugar Beet Research, II, p139, Brussels: IIBR, 1987.
- Li, Yu, Han, Xing, Liu & Chen, Ping Tu Hsu Pao 14: 165, 1998.
- Li, Yu, Han, Xing, Liu & Koenig, Chinese Journal of Biotechnology 15: 461, 1999.
- Lindsten, Bulletin OEPP/EPPO Bulletein 19: 531, 1989.
- Mahmood & Rush, Plant Disease 83: 521, 1999.
- Mannerlöf, Lennerfors & Tenning, Euphytica 90: 293, 1996.
- Martine & Whitney, Plant Disease 74: 31, 1990.
- Miyanishi, Kusume, Saito & Tamada, Archives of Virology 144: 879, 1999.
- Morales, Ward, Castano, Arroyave, Lozano and Adams, European Journal of Plant Pathology 105: 643, 1999.
- Niesbach-Klosgen, Guilley, Jonard & Richards, Virology 178: 52, 1990.
- Paul, Henken & Alderlieste, Netherlands Journal of Plant Pathology 98: 65, 1992.
- Pelsy & Merdinoglu, Plant Breeding 115: 371, 1996.
- Poggi-Pollini & Giunchedi, Phytopathologica Mediterranea 28: 16, 1989.
- Putz, Journal of General Virology 35: 397, 1977.
- Putz & Vuittenez, Journal of General Virology 50: 201, 1980.
- Putz, Wurtz, Merdinoglu, Lemaire & Valentin, in Viruses with Fungal Vectors, p. 83, J. I. Cooper & M. J. C. Asher, eds, Wellesbourne: Association of Applied Biologists, 1988.
- Richard-Molard, Span 28: 92, 1985.
- Richards & Tamada, Annual Review of Phytopathology 30: 291, 1992.
- Richards, Jonard, Guilley, Ziegler & Putz, Journal of General Virology 66: 345, 1985.
- Rush & Heidel, Plant Disease 79: 868, 1995.
- Rush, French & Heidel, Phytopathology 84: 1366, 1994.
- Russo, Martelli & di Franco, Physiological Plant Pathology 19: 237, 1981.
- Rysanek, Stocky, Haeberle & Putz, Agronomie 12: 651, 1992.
- Saito, Kiguchi, Kusume & Tamada, Archives of Virology 141: 2163, 1996.
- Saito, Kiguchi & Tamada, Bulletin of the Research Institute for Bioresources, Okayama University 5: 79, 1997.
- Salle, Le Coz & Tuquet, Physiologie Vegetale 24: 73, 1986.
- Schlosser, in Viruses with Fungal Vectors, p. 281, J. I. Cooper & M. J. C. Asher, eds, Wellesbourne: Association of Applied Biologists, 1988.
- Schmitt, Balmori, Jonard, Richards & Guilley, Proceedings of the National Academy of Sciences, USA 89: 5715, 1992.
- Scholten, Paul, Peters, van Lent & Goldbach, Archives of Virology 136: 349, 1994.
- Scholten, Jansen, Paul, Keizer, de Bock & Lange, Euphytica 91: 331, 1996.
- Steven, Trus, Putz & Wurtz, Virology 113: 428, 1981.
- Tamada, CMI/AAB Descriptions of Plant Viruses 144: 1975.
- Tamada, in Encyclopedia of Virology, 2nd Edition, p. 154, A. Granoff & R. G. Webster, eds, London: Academic Press, 1999.
- Tamada & Abe, Journal of General Virology 70: 3391, 1989.
- Tamada & Baba, Annals of the Phytopathological Society of Japan 39: 325, 1973.
- Tamada & Kusume, Journal of General Virology 72: 1497, 1991.
- Tamada, Shirako, Abe, Saito, Kiguchi & Harada, Journal of General Virology 70: 3399, 1989.
- Tamada, Schmitt, Saito, Guilley, Richards & Jonard, Journal of General Virology 77: 1359, 1996a.
- Tamada, Kusume, Uchino, Kiguchi & Saito, in Proceedings, 3rd Symposium of the International Working Group on Plant Viruses with Fungal Vectors, p.49, Denver: American Society of Sugar Beet Technologists, 1996b.
- Tamada, Uchino, Kusume & Saito, Phytopathology 89: 1000, 1999.
- Torrance & Mayo, Archives of Virology 142: 435, 1997.
- Torrance, Pead & Buxton, Annals of Applied Biology 113: 519, 1988.
- Tuitert, Netherlands Journal of Plant Pathology 96: 331, 1990.
- Tuitert & Hofmeester, Netherlands Journal of Plant Pathology 98: 343, 1992.
- Tuitert & Hofmeester, European Journal of Plant Pathology 100: 19, 1994.
- Tuitert, Musters-Van Oorschot & Heijbroek, European Journal of Plant Pathology 100: 201, 1994.
- Whitney, Plant Disease 73: 287, 1989.
- Wisler, Liu & Duffus, Plant Disease 78: 995, 1994.
- Yao, Liu, Yu, Cai, Ao & Yang, Chinese Journal of Biotechnology 9: 131, 1993.
- Yu, Li, Yang & Liu, Chinese Journal of Biotechnology 11: 119, 1995.
- Yu, Han, Yang, Li & Liu, Acta Microbiologica Sinica 37: 7, 1997.
- Ziegler, Richards, Guilley, Jonard & Putz, Journal of General Virology 66: 2079, 1985.