Details of DPV and References
DPV NO: 399 July 2003
Family: Potyviridae
Genus: Potyvirus
Species: Lettuce mosaic virus | Acronym: LMV
This is a revised version of DPV 9
Lettuce mosaic virus
Olivier Le Gall IBVM, INRA Bordeaux-Aquitaine, BP 81, F-33883 Villenave d'Ornon Cedex, France
Contents
- Introduction
- Main Diseases
- Geographical Distribution
- Host Range and Symptomatology
- Strains
- Transmission by Vectors
- Transmission through Seed
- Transmission by Grafting
- Transmission by Dodder
- Serology
- Nucleic Acid Hybridization
- Relationships
- Stability in Sap
- Purification
- Properties of Particles
- Particle Structure
- Particle Composition
- Properties of Infective Nucleic Acid
- Molecular Structure
- Genome Properties
- Satellite
- Relations with Cells and Tissues
- Ecology and Control
- Notes
- Acknowledgements
- Figures
- References
Introduction
Lettuce mosaic disease was first described in Florida (Jagger, 1921). A comprehensive review of the properties of the virus was published by Dinant & Lot (1992).
- Selected synonyms:
- LMV was named Lactuca virus 1 (Smith, 1937) and Marmor lactucae (Holmes, 1939) in early descriptions but was soon known under its current name, Lettuce mosaic virus (Ainsworth & Ogilvie, 1939).
Brief description
LMV is a virus with flexuous filamentous particles approximately 750 x 13 nm. It is sap-transmissible to a wide range of species, often seed-borne in lettuce and transmitted by several aphid species in the non-persistent manner. It has a world-wide distribution and a relatively broad host range among dicotyledoneous plants, including many Asteraceae. LMV can cause economic losses in cultivated Asteraceae, especially lettuce (Lactuca sativa) and ornamentals.
Main Diseases
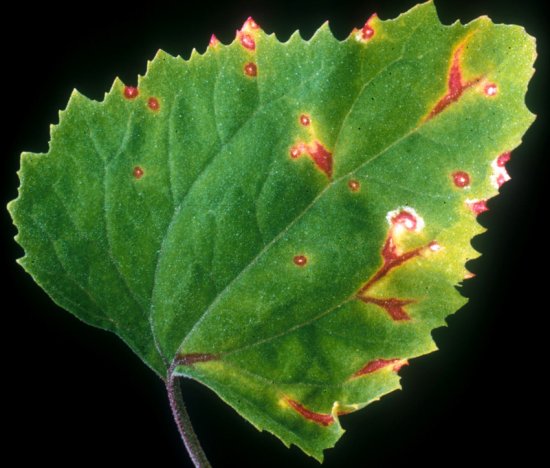
Causes various mosaic and mottle symptoms in all types of lettuce (Fig. 1, Fig. 2). Natural infection has also been reported in endive/escarole (Cichorium endiva), pea (Pisum sativum), chickpea (Cicer arietinum), mustard (Sinapis arvensis), spinach (Spinacia oleracea), safflower (Carthamus tinctorius) and various ornamentals (Osteospermum spp., Gazania spp., Bupleurum falcatum, etc).
Geographical Distribution
World-wide where lettuce is grown (Krause-Sakate et al., 2002).
Host Range and Symptomatology
Host range is wide (Costa & Duffus, 1958) and covers several dicotyledoneous families. Weeds and ornamentals can act as local reservoirs for lettuce crops. In particular many species within the family Asteraceae are susceptible to LMV, including cultivated and ornamental species such as lettuce (Lactuca sativa), endive/escarole (Cichorium endiva), safflower (Carthamus tinctorius), starthistle (Centaurea solstitialis), osteospermum and gazania. In addition, several species within the families Brassicaceae, Cucurbitaceae, Fabaceae, Solanaceae and Chenopodiaceae are natural or experimental hosts of LMV.
- Diagnostic species
Lactuca sativa (lettuce). Symptoms are variable but usually consist of vein clearing in the younger leaves and mosaic in the older ones. Veinal necrosis occurs with some isolate/cultivar combinations, as well as lethal wilting. Plants typically fail to 'heart' and inner leaves remain dwarfed and rosetted (Fig. 1, Fig. 2).
Chenopodium amaranticolor. Pale green or chlorotic local lesions (usually with reddish margins) after 8-10 days (Fig. 3).
C. quinoa. Pale green local lesions. Conspicuous, systemic yellow vein-net symptoms with twisting and stunting of apical leaves (Fig. 4).
Gomphrena globosa. Whitish, local necrotic dots (4-7 days) enlarging into red-rimmed lesions.
Pisum sativum (pea). Systemic infection produces within a few days vein clearing and mosaic symptoms similar to those of Pea seed-borne mosaic virus.
Nicotiana benthamiana. Systemic infection results in mosaic symptoms.
Osteospermum ecklonis. Leaf symptoms are often a more or less severe mosaic with distortion. Flower discolorations are not frequent but sometimes observed (Fig. 5).
- Non host species
Species described as non-hosts include sunflower (Helianthus annuus), tomato (Lycopersicon esculentum), cabbage (Brassica oleracea), cucumber (Cucumis sativus), carrot (Daucus carota), Vigna spp. and monocots (Provvidenti & Schroeder, 1972). The host status depends on the cultivars or accessions in several species such as bean (Phaseolus vulgaris), pumpkin (Cucurbita maxima), Arabidopsis thaliana, etc, raising the doubt that the diversity of some species described as non-hosts might simply have been too little explored.
- Propagation species
Lettuce plants 10-15 days after inoculation are suitable sources of virus for purification. Pea, Chenopodium quinoa, Nicotiana benthamiana, N. clevelandii and safflower have also been used for virus purification but lettuce seems to provide the best yields (Dinant & Lot, 1992).
- Assay species
Chenopodium amaranticolor, C. quinoa and Gomphrena globosa are suitable local lesion hosts. Chenopodium quinoa has also been used for seed indexing (Pelet, 1965).
Seed transmission and vector transmission can be assayed in lettuce.
Non-hosts such as Vigna spp. allow discrimination of possible co-infection with other viruses which produce local symptoms and/or systemic infection (e.g. Cucumber mosaic virus).
Lettuce cultivars differing in their resistance genotype can be used to distinguish virus strains.
Strains
Strains and pathotypes have been described on the basis of symptoms in various hosts including differential lettuce cultivars (McLean & Kinsley, 1963; Pink et al., 1992). However, while this classification is clearly supported by biological observations, it is only very partially so by sequence clustering analysis (Krause-Sakate et al., 2002). This is indicative of functional convergence.
Sequence analysis revealed that LMV isolates could be clustered in three groups: a single isolate from Yemen, a group from the Balkans (Greece and Croatia) and a third group with very diverse geographical origins (including the Middle East and Greece). Within this third group, two large sub-clusters of isolates, collectively named "LMV-Common" and "LMV-moST" (for mo1-breaking, Seed Transmitted), contain all known seedborne isolates. Given the propagation of lettuce through seeds, this probably explains why LMV-Common and LMV-moST account for the vast majority of lettuce isolates.
The two recessive resistance alleles of the mo1 gene, used to protect lettuce world-wide, are effective against LMV-Common but not against other types of isolates. Outbreaks of resistance-breaking isolates usually remain localised because they are caused by non-seedborne isolates. LMV-moST is a remarkable exception and these resistance-breaking seedborne isolates, first discovered in the early 1990's (Dinant & Lot, 1992), have now been spread to several continents, probably through commercial exchange of contaminated seeds (Krause-Sakate et al., 2002).
Transmission by Vectors
LMV is transmitted in a non-persistent manner but with varying efficiencies by several species of aphids (Acyrthosiphon scariolae, Acyrthosiphon pisum, Aphis gossypii, Myzus persicae, Macrosiphum euphorbiae, Nasonovia ribisnigri, Rhopalosiphum pseudobrassicae, etc.). M. persicae is a particularly efficient vector; all instars of M. persicae transmit LMV but alates are less efficient than apterae. Transmission efficiency increases with increasing periods of fasting (5-240 min), but decreases with increasing acquisition access time from 5 to 120 min (Sylvester, 1955).
Transmission through Seed
LMV-Common and LMV-moST are seedborne in lettuce while other isolates are apparently not (Newhall, 1923; Krause-Sakate et al., 2002). LMV-Common is not transmitted from lettuce cultivars carrying the mo1 resistance alleles even in those cases where symptomless virus accumulation occurs (Dinant & Lot, 1992). Seed transmission is probably the major factor in the dissemination of the mosaic disease in lettuce crops since it provides the primary inoculum for further spread by aphids (Fig. 6).
Some 3-15% of the seeds collected from an infected plant give rise to infected seedlings, depending on the time of infection of the mother plant, the variety and the environmental conditions (Dinant & Lot, 1992). Seed transmission also occurs in the wild prickly lettuce Lactuca serriola (van Hoof, 1959).
Seed transmission has been reported to occur through pollen but mostly maternally (Ryder, 1964). Because more recent ultramicroscopy studies could not detect virus particles or other viral ultrastructures in unfertilized ovules (Hunter & Bowyer, 1994), infection of seed seems to occur post-fertilization. The production of infected pollen follows early invasion of the floral meristem (Hunter & Bowyer, 1997).
Transmission by Dodder
No reports.
Serology
The virus is a relatively good immunogen and high quality antisera have been obtained and used in serological detection techniques (gel diffusion, ELISA, ISEM, RIA, Immunocapture-based RT-PCR). Monoclonal antibodies have also been obtained.
Relationships
Monoclonal antibodies obtained against LMV virions had differential reactivities when tested against several isolates. However LMV-Common and LMV-moST are not separated by available monoclonal antibodies, and the amino-terminal extension of their capsid protein, a domain which is both especially immunogenic and variable in potyviruses, is in fact identical in sequence between these two strains (Krause-Sakate et al., 2002).
The sequence of the hypervariable NIb-CP junction (Krause-Sakate et al., 2002) is 70-75% identical between isolates of the three main groups (Yemen, Balkan and "main"). It is 87-92% identical between isolates belonging to different branches within the major cluster, and it is more than 92% identical within a given branch. LMV-moST isolates are less divergent in sequence from each other than are LMV-Common isolates.
In cross-protection tests, some LMV isolates protected lettuce against other more virulent isolates (McLean & Kinsley, 1963).
Various techniques (microprecipitation, precipitin, gel diffusion, ISEM, ELISA using monoclonal antibodies, etc) indicated that LMV is distantly related serologically to several other potyviruses such as Soybean mosaic virus, Maize dwarf mosaic virus, Sugarcane mosaic virus, Turnip mosaic virus, Bean yellow mosaic virus, Pea mosaic virus, Clover yellow vein virus, and possibly Potato virus Y (Brandes & Bercks, 1965; Dinant & Lot, 1992). Other potyviruses were found not to be serologically related to LMV (Bidens mottle virus, Iris mosaic virus, Parsnip mosaic virus, Bean common mosaic virus).
Stability in Sap
In lettuce sap, the thermal inactivation point (10 min) is 55-60°C, and the dilution end-point is 10-2-10-3 (Tomlinson, 1962b; Kuida et al., 1977). Infectivity is retained at 20°C for 1-4 days (Kuida et al., 1977; Qian et al., 1996). Infectivity of purified virus preparations is considerably stabilized by adding 0.5% sodium sulphite (Ainsworth & Ogilvie, 1939) or 0.1% thioglycollic acid (Tomlinson, 1964). Dessicated lettuce tissues retain infectivity for months or years and provide a simple way to store isolates.
Purification
Many purification methods use buffers with high ionic strength (0.1-0.5M sodium citrate, sodium borate, potassium phosphate or Tris) and high pH (8-9) in the first steps, together with organic solvents (butanol, freon 113, chloroform/carbon tetrachloride). After homogenization and elimination of the largest debris, differential centrifugation is usually preferred to polyethylene glycol precipitations (Dinant & Lot, 1992).
In a typical purification protocol (Dinant & Lot, 1992), infected lettuce leaves are homogenized in 0.3M K2HPO4 containing DIECA and 2-mercaptoethanol, an equal volume of freon 113 is added and the mixture is centrifuged at low speed. Triton X-100 (1%) is added to the aqueous phase. The mixture is gently stirred in the cold for 20 minutes and subjected to differential centrifugation (high speed concentration, low speed clarification, high speed concentration). A final isopycnic centrifugation in caesium sulphate yields 8 to 18 mg virus per kilogram of lettuce leaves.
Properties of Particles
The sedimentation coefficient is 143 S. The buoyant density is 1.33 g.cm-3 in caesium chloride at 20°C (Huttinga & Mosch, 1974).
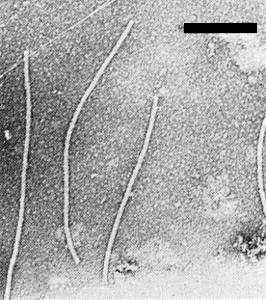
Particle Structure
LMV has typical potyvirus particles. Particles are flexuous filaments approximately 750 x 13 nm (Fig. 7) (Moghal & Francki, 1981). They do not appear to be penetrated by phosphotungstate.
Particle Composition
The particles are exclusively made of a genomic RNA encapsidated in a single type of capsid protein subunit. The genome is a single-stranded RNA of 10080 nucleotides, with a polyadenylated 3' end and a virus-encoded protein (VPg) covalently bound to the 5' end. The molecular weight of the capsid protein subunit has been estimated electrophoretically to be 34 kDa (Huttinga & Mosch, 1974) and calculated from the nucleotide sequence to be 31 kDa (Dinant et al., 1991).
Genome Properties
The genetic organisation of the four completely sequenced LMV isolates (EMBL accession numbers: X97704 and AJ306288 for LMV-Common, AJ278854 for LMV-moST and X97705 for another type of LMV) is typical for potyviruses (Revers et al., 1997; Krause-Sakate et al., 2002). The RNA genome is 10080 nucleotides (excluding the 3' poly-A) in all four complete sequences determined so far and encodes a single large polyprotein (3255 amino-acids) processed by three virus-encoded proteinases (P1Pro, HCPro and NIaPro). The capsid protein domain maps to the carboxy-terminus of the polyprotein.
Relations with Cells and Tissues
Electron microscopy of cells infected by LMV shows the presence of various types of cytoplasmic inclusions and cytopathogenic effects. Pinwheels and laminated cytoplasmic aggregates are consistently observed. The cell structures appear unchanged, except for a reduction in size of the chloroplasts, the presence of plastoglobules in chloroplasts, and enlarged mitochondria in Chenopodium quinoa cells but not lettuce cells (Saric & Wrischer, 1977). Extensive electron microscopy studies of floral tissues indicate that LMV particles and cytoplasmic pinwheel and bundle inclusions are present in all types of cells investigated but were not observed in the embryo sac (Hunter & Bowyer, 1994; Hunter & Bowyer, 1997).
Ecology and Control
Spread occurs through the use of infected seeds (Fig. 6) and from neighbouring infected lettuce, ornamentals or weeds through aphids. The main methods to control the disease are the use of virus-free seeds (Tomlinson, 1962a), and elementary sanitary measures (keeping lettuce nurseries away from fields in which crops are grown, etc). Alleles of the recessive resistance gene mo1 (Bannerot et al., 1969; Ryder, 1970), which encodes the translation initiation factor 4E (Nicaise et al., 2003), are successfully used to control LMV-Common but not other isolates, including the seedborne LMV-moST. Other sources of resistance have been identified in wild relatives of lettuce. Expression of the capsid protein from a transgene protects lettuce against LMV infection (Dinant et al., 1997).
Notes
While LMV was considered to be efficiently controlled by the use of mo1 resistance alleles, the spread in the last decade of mo1-breaking isolates that are transmitted through seed, and which have recently been assigned to a single strain, LMV-moST, is a cause of concern for lettuce production world-wide. We have recently found a recombinant LMV isolate resulting from a natural exchange between LMV-moST and LMV-Common in a field where both strains occurred (Krause-Sakate et al., in press).
A specific effort to avoid the presence of LMV-moST in lettuce (dissemination of virus-free seeds, specific detection tools, identification and control of possible reservoirs, identification of genetic resistance sources, etc) must be achieved and will best be achieved collectively.
Figures

Lettuce (Butterhead cv. Trocadéro) with systemic symptoms and defective heading caused by LMV. Photo: INRA Bordeaux, France.

Mosaic symptoms caused by LMV in a field-grown butterhead-type lettuce in France (photo: INRA Avignon, France).
Reddish local lesions caused by LMV in a rub-inoculated leaf of Chenopodium amaranticolor. Photo: INRA Bordeaux, France.

Lettuce seedlings growing in a nursery before transplantation to the field in Tunisia. LMV-infected plantlets growing from infected seeds, such as the one on the right, provide the primary inoculum for aphid-mediated spread of the mosaic disease through the nursery and in lettuce crops. Photo: INRA Bordeaux, France.
References list for DPV: Lettuce mosaic virus (399)
- Ainsworth & Ogilvie, Annals of Applied Biology 26: 279, 1939.
- Bannerot, Boulidard, Marrou & Duteil, Annales de Phytopathologie 1: 219, 1969.
- Brandes & Bercks, Advances in Virus Research 11: 1, 1965.
- Costa & Duffus, Plant Disease Reporter 42: 583, 1958.
- Dinant & Lot, Plant Pathology 41: 528, 1992.
- Dinant, Lot, Albouy, Kuziak, Meyer & Astier-Manifacier, Archives of Virology 116: 235, 1991.
- Dinant, Maisonneuve, Albouy, Chupeau, Chupeau, Bellec, Gaudefroy, Kusiak, Souche, Robaglia & Lot, Molecular Breeding 3: 75, 1997.
- Holmes, Handbook of phytopathogenic viruses, p. 221, Minneapolis: Burgess Publishing Company, 1939.
- Hunter & Bowyer, Journal of Phytopathology 140: 11, 1994.
- Hunter & Bowyer, Journal of Phytopathology 145: 521, 1997.
- Huttinga & Mosch, Netherlands Journal of Plant Pathology 80: 19, 1974.
- Jagger, Journal of Agricultural Research 20: 737, 1921.
- Krause-Sakate, Fakhfakh, Peypelut, Pavan, Zerbini, Marrakchi, Candresse & Le Gall, Archives of Virology, 149: 191, 2004.
- Krause-Sakate, Le Gall, Fakhfakh, Peypelut, Marrakchi, Varveri, Pavan, Souche, Lot, Zerbini & Candresse, Phytopathology 92: 563, 2002.
- Kuida, Inouye & Inouye, Nogaku Kenkyu 56: 33, 1977.
- McLean & Kinsley, Phytopathology 52: 403, 1963.
- Moghal & Francki, Virology 112: 210, 1981.
- Newhall, Phytopathology 13: 104, 1923.
- Nicaise, German-Retana, Sanjuan, Dubrana, Mazier, Maisonneuve, Candresse, Caranta & Le Gall, Plant Physiology 132: 1272, 2003.
- Pelet, Revue Horticole Suisse 38: 7, 1965.
- Pelet, Revue Horticole Suisse 44: 172, 1971.
- Pink, Kostova & Walkey, Plant Pathology 41: 5, 1992.
- Provvidenti & Schroeder, Plant Disease Reporter 56: 281, 1972.
- Qian, Luo & Gong, Journal of Zhejiang Agricultural University 22: 304, 1996.
- Revers, Yang, Walter, Souche, Lot, Le Gall, Candresse & Dunez, Virus Research 47: 167, 1997.
- Ryder, Plant Disease Reporter 48: 522, 1964.
- Ryder, Journal of the American Society for Horticultural Sciences 95: 378, 1970.
- Saric & Wrischer, Phytopathologische Zeitschrift 90: 27, 1977.
- Smith, A handbook of plant virus diseases, p. 652, Boston: Little, Brown & Co, 1937
- Sylvester, Phytopathology 45: 357, 1955.
- Tomlinson, Plant Pathology 11: 61, 1962a.
- Tomlinson, Nature 193: 299, 1962b.
- Tomlinson, Annals of Applied Biology 53: 95, 1964.
- van Hoof, Tijdschrift over Plantenziekten 6: 44, 1959.