Details of DPV and References
DPV NO: 403 January 2004
Family: Tombusviridae
Genus: Aureusvirus
Species: Pothos latent virus | Acronym: PoLV
Pothos latent virus
P. Lava Kumar International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Patancheru 502 324, AP, India
G. P. Martelli Dipartimento di Protezione delle Piante e Microbiologia Applicata, Università degli Studi and Istituto di Virologia Vegetale del CNR, sezione di Bari, 70126 Bari, Italy
Contents
- Introduction
- Main Diseases
- Geographical Distribution
- Host Range and Symptomatology
- Strains
- Transmission by Vectors
- Transmission through Seed
- Transmission by Grafting
- Transmission by Dodder
- Serology
- Nucleic Acid Hybridization
- Relationships
- Stability in Sap
- Purification
- Properties of Particles
- Particle Structure
- Particle Composition
- Properties of Infective Nucleic Acid
- Molecular Structure
- Genome Properties
- Satellite
- Relations with Cells and Tissues
- Ecology and Control
- Notes
- Acknowledgements
- Figures
- References
Introduction
First described by Sabanadzovic et al. (1995).
- Synonym:
- PoLV-PP [isolate from pigeonpea (Cajanus cajan) (Kumar et al., 2001)]
Brief description:
An isometric virus c. 30 nm in diameter, containing a single-stranded molecule of positive sense RNA. This virus was first isolated from hydroponic cultures of pothos (Scindapsus aureus). The virus causes symptomless infections in the natural hosts reported. The virus is very stable and is readily sap-transmissible to several herbaceous species. It has no known vector, but is transmitted through soil by direct ingress through roots.
Main Diseases
Symptomless in pothos and in pigeonpea.
Geographical Distribution
Reported only from southern Italy and the central part of India (Sabanadzovic et al., 1995; Kumar et al. 2001).
Host Range and Symptomatology
Pothos is the only known natural systemic host and infection is symptomless. In Italy, the virus was detected in pothos plants in hydroponic culture in glasshouses, but not in those from outside (Sabanadzovic et al., 1995). In pigeonpea, the virus was detected in concentrated leaf extracts of field-collected samples from some locations in India (Kumar et al., 2001).
The experimental host range is moderately wide (Kumar et al., 2001; Sabanadzovic et al., 1995), infecting several species in 6 botanical families. Several infected plants develop necrotic symptoms in inoculated leaves but others are infected symptomless. Systemic infection occurs in only a few species, and some show systemic mosaic and mottling followed by necrosis and death within 2 - 3 weeks post inoculation.
- Diagnostic species:
- Chenopodium quinoa. Pin-point local necrotic lesions that expand to form large necrotic patches (Fig. 1).
- Gomphrena globosa. Large necrotic local lesions with purple margins. Veinal purpling often limited to the mid-vein portion of a few leaves when infection is systemic (Fig. 2).
- Nicotiana clevelandii. Systemic necrosis (Fig. 4)
- Nicotiana hispens. Systemic necrosis resulting in death of apical shoots.
- Ocimum basilicum. Black necrotic local lesions.
- Spinacia oleraceae. Large round sunken local necrotic lesions.
- Propagation species:
- Nicotiana clevelandii, N. benthamiana and N. hispens are useful for virus propagation and G. globosa for long-term maintenance of virus cultures.
- Assay species:
- Chenopodium quinoa, C. amaranticolor and C. murale are useful local lesion hosts. N. benthamiana is useful for assaying systemic infection.
- Non-hosts:
- Cucurbita pepo, C. sativus, Datura stramonium, Nicotiana glutinosa, N. tabacum cv. White Burley, Vicia faba cvs. Minden, The Sutton, and Vigna unguiculata.
Strains
None reported; an isolate from pigeonpea (PoLV-PP) has a very close sequence homology to the type isolate (Kumar et al., 2001).
Transmission by Vectors
No vector reported. Natural transmission occurs through the soil without the involvement of a vector. Virus particles exuding through roots can serve as sources of infection for other plants (Sabanadzovic et al., 1995). Soil-borne transmission through direct ingress of virus through roots was demonstrated (Kumar et al., 2001).
Transmission through Seed
No information.
Transmission by Dodder
No information.
Serology
The virus is a good immunogen producing antiserum with gel double-diffusion titres of 1/1024 and polyclonal antiserum gives a single precipitin line in immunodiffusion tests in 1% agarose gel. In immuno-sorbent electron microscopy (ISEM) assays, antibodies uniformly decorate the virus particles. Virus can be detected by a wide range of serological assays, most commonly using direct or indirect ELISA formats. Agar gel double-diffusion tests are commonly used for studying serological relationships among the members of the family Tombusviridae (Sabanadzovic et al., 1995; Kumar et al., 2001).
Relationships
Isolates from pothos and pigeonpea are serologically indistinguishable and share 93% sequence homology (Table 1). The relationship of PoLV with other members of the Family Tombusviridae is established by serological cross reactivity and by sequence comparison. Serologically distantly related to Eggplant mottled crinkle virus, Sikte waterborne virus and Lato river virus of the genus Tombusvirus; and Ahlum waterborne virus and Galinsoga mosaic virus of the genus Carmovirus (Sabanadzovic et al., 1995). The genome organisation and expression strategy is identical to that of species in the genus Tombusvirus. However, the size of ORF3 and ORF4 of tombusviruses and aureusviruses are significantly different (Rubino et al., 1995). The amino acid sequences of proteins encoded by the ORFs 1, 4 and 5 are distantly related to the comparable tombusvirus proteins. PoLV does not support replication of defective interfering RNAs of tombusviruses indicating that its replicase may have different recognition sequences from those of tombusviruses (Rubino and Russo, 1997).
Table 1: Pair-wise comparison of PoLV, PoLV-PP and CLSV amino acid sequences deduced from their respective ORFs.
| Virus pair | % amino acid sequence identity | ||||
| ORF 1 | ORF 2 | ORF 3 | ORF 4 | ORF 5 | |
| PoLV/CLSV | 63 | 87 | 37 | 78 | 60 |
| PoLV/PoLV-PP | 96 | 96 | 92 | 97 | 98 |
| PoLV-PP/CLSV | 64 | 80 | 39 | 77 | 59 |
Stability in Sap
The virus is stable and infectivity is retained for long periods. In Nicotiana clevelandii leaves stored at 4 °C and -20 °C, virus infectivity was retained for up to 90 days, the longest period tested. In crude plant sap extracts, infectivity survived storage for more than 32 days at room temperature, 4 °C and -20 °C. Virus infectivity was retained for more than one year (the longest period tested) in freeze-dried leaf tissue (Kumar et al., 2001). The thermal inactivation point is above 80 °C. The virus is resistant to organic solvents, but is rapidly disrupted in the presence of detergents such as SDS (Kumar et al., 2001).
Purification
Because of the high stability of virus particles and its occurrence in high titres in plants, it can be purified easily from infected plants in yields of up to 40-80 mg/kg of tissue (Sabanadzovic et al., 1995; Kumar et al., 2001). The following method is satisfactory. Extract infected leaves of N. clevelandii or N. benthamiana in 50 mM phosphate buffer pH 7, containing 0.2% b-mercaptoethanol (or a-monothioglycerol) at 2 ml buffer/g leaf. Squeeze the extract through a single layer of muslin cloth and clarify it with an equal volume of chloroform. Centrifuge the emulsion at 12,000 g for 15 min, collect the aqueous phase and concentrate it by centrifuging at 278,000g for 60 min. Resuspend the pellets in 1-3 ml 50 mM phosphate buffer and layer on a 10-40% linear sucrose density gradient and centrifuge at 160,000g for 2 h. Collect the light-scattering zone, dilute it with 2.5 volumes of 50 mM phosphate buffer and concentrate virus particles by centrifuging at 45,000 rpm for 90 min. Resuspend the virus-containing pellets in a minimal volume of 50 mM phosphate buffer.
Properties of Particles
Purified preparations of virus particles sediment as a single major component in sucrose density gradients and have a buoyant density in CsCl and Cs2SO4 of 1.36 g/cm3 and 1.27 g/cm3, respectively (Kumar et al. 2001).
A260/280: 1.65; Amax258 nm; Amin 242 nm
Purified virus particles are infective.
Particle Structure
Particles are isometric with a knobbly surface and are c. 30 nm in diameter (Fig. 5). Particles are penetrated by 2% phosphotungstic acid (PTA), but not by uranyl acetate (Kumar et al. 2001). Amino acid sequence alignments suggests that the coat protein subunits may be made up of three structural domains, the N-terminal domain, the shell domain and the C-terminal protruding domain (Rubino and Russo, 1997).
Particle Composition
Nucleic acid:
Virus particles contain a single molecule of linear ssRNA of 4415 nts. Genomic RNA is positive sense, uncapped and not polyadenylated. RNA constitutes 17% of the particle weight. Virions sometimes encapsidate two subgenomic (sg) RNAs of size 2 and 0.8 kb. Three dsRNA species corresponding to the full-size genomic RNA and two sub-genomic RNAs are found in infected plants. Satellite or defective interfering RNAs were not detected in naturally occurring isolates (Sabanadzovic et al., 1995; Rubino and Russo, 1997; Kumar et al., 2001).
Protein:
Virions contain a single coat protein (CP) with a Mol. Wt of 40,000 estimated from nucleotide sequence data. The CP resolves as a single band in denaturing polyacrylamide gels with an estimated Mol. Wt of 42-46,000. Virions are predicted to contain 180 copies of the CP subunit (Rubino and Russo, 1997).
Lipid:
No information.
Other components:
No information.
Genome Properties
Two isolates are sequenced completely [accession numbers X87115 (PoLV); AJ243370 (PoLV-PP)] and are 93% identical with a high degree of amino acid sequence identity (Table 1; Kumar et al., 2001). The virus genome contains 5 ORFs (Rubino et al., 1995; Rubino and Russo, 1997). The replication strategy includes read-through and sgRNA production. ORFs 1 and 2 are expressed directly from the genomic RNA, whilst the remaining three ORFs downstream of ORF2 are expressed through two 3' co-terminal sgRNAs of size 2 and 0.8 kb (Rubino and Russo, 1997). ORF 1 encodes a putative 25 kDa protein of unknown function. ORF2 is a read-through of an amber stop codon of ORF1 and is expressed as an 84 kDa protein, which is considered to have a replicase function. ORF 3 encodes the coat protein of 40 kDa. ORF 4 encodes a 27 kDa protein and ORF 5, completely nested but in a different reading frame of ORF 4, expresses a 14 kDa protein (Fig. 6). Mutagenesis studies identified the 27 kDa product expressed by ORF 4 as the protein involved in cell-to-cell and long distance movement, and the 14 kDa product of ORF 5 as a component for symptom severity (Rubino and Russo, 1997).
Relations with Cells and Tissues
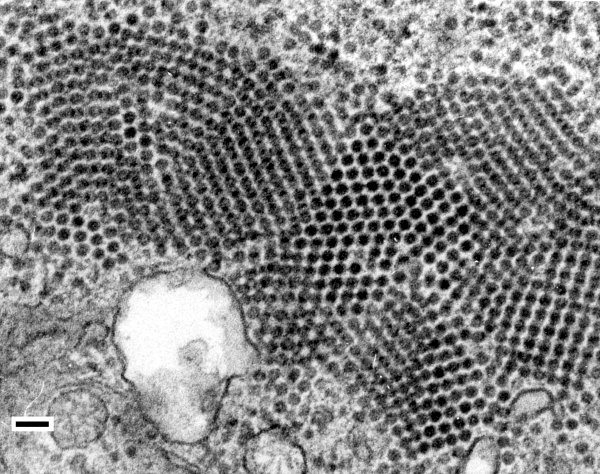
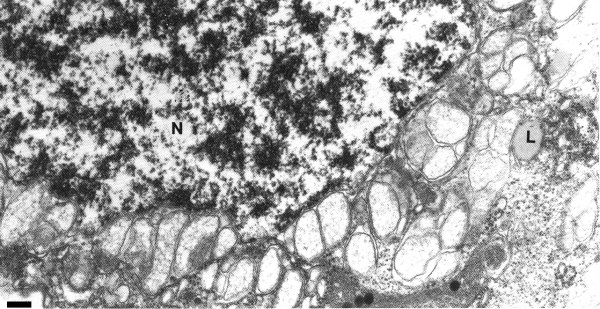
Virus is found in all types of cells including conducting tissues (Sabanadzovic et al., 1995; Russo et al., 1997). Virus particles assemble and accumulate in the cytoplasm of infected cells. Virions are found throughout the cytoplasm, but they also occur in discrete aggregates and in regularly arranged crystalline lattices (Fig. 7). Excessive vesiculation of the nuclear envelope and single-membrane vesiculated bodies protruding into the vacuoles is a prominent cytopathological feature (Fig. 8). Replication is thought to occur in the cytoplasm within the nucleus-derived globose vesicles surrounded by unit membranes (Russo et al., 1997).
Ecology and Control
The virus was first identified as a surface contaminant on rooted cuttings of Vitis rupestris (a non-host) in hydroponic cultures (Sabanadzovic et al., 1995). Subsequent characterisation identified pothos as the virus source for plants grown in hydroponic cultures. The virus was also isolated from concentrated leaf extracts of field-collected pigeonpea but it is not systemic in this host, although it is retained on inoculated leaves for long periods (Kumar et al., 2001). Based on these observations, it is assumed that on pigeonpea, the virus may be a surface contaminant (Kumar et al., 2001). Limited surveys for the natural occurrence of the virus showed that it is limited to plants grown in hydroponic cultures in the southern Italy. It was not detected in pothos samples collected from outside glasshouses (Sabanadzovic et al., 1995). The true source of infection on pigeonpea is not known (Kumar et al., 2001). Like several tombusviruses, owing to its high stability and infectivity in vitro, and its ability to spread and infect plants through roots without a vector, the virus probably survives and spreads in soil and soil water (Koenig, 1986; Sabanadzovic et al., 1995; Kumar et al., 2001).
Notes
The virus shares many similarities with tombusviruses. However, the expression products of ORFs 1, 4 and 5 are only distantly related to comparable tombusvirus proteins. A new genus, Aureusvirus, within the family Tombusviridae was therefore established, with PoLV as the type member (Martelli et al., 1998). Cucumber leaf spot virus (CLSV), transmitted by the chytrid fungus Olpidium bornovanus (Miller et al., 1997; Martelli et al., 1998; Reade et al., 2003) was placed in this genus based on the homology with PoLV of the polymerase domain sequence (ORF 2) and in the size and sequence homology of the products of ORFs1, 2 and 4. Apart from ORFs 2 and 4, the amino acid sequence of these two viruses is significantly different (Table 1).
Figures

Nicotiana benthamiana plants showing progressive systemic wilting and death, from left to right, 2, 6, 12 and 16 days after inoculation.
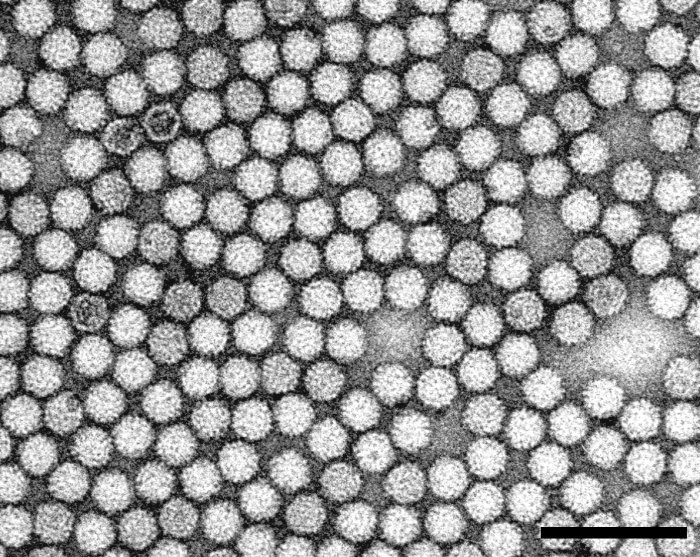
Electron micrograph of a purified virus particle preparation negatively stained with 2% phosphotungstic acid. Bar = 100 nm.

Genome structure and organisation. The RNA is indicated with lines and the ORFs as blocks. The 4.4 kb genomic RNA encodes for 5 proteins. ORF 1 and ORF 2 are encoded by direct expression from genomic RNA, whereas ORFs 3, 4 and 5 are expressed through two sub-genomic (sg) 3' co-terminal RNAs. ORF 1 encodes for a 25 kDa protein and ORF 2, a read-through of an amber stop codon (arrow) of ORF 1 that encodes an 84 kDa replicase protein. ORF 3 expressed from a 2 kb sgRNA-1 encodes the 40 kDa coat protein. ORF 4 is expressed from a 0.8 kb sgRNA-2 that encodes a 27 kDa movement protein. ORF 5, that is completely nested in ORF 4 but in a different reading frame, is expressed from the sgRNA-2 and is responsible for symptom severity.
References list for DPV: Pothos latent virus (403)
- Koenig, Advances in Virus Research 31: 321, 1986.
- Kumar, Jones, Sreenivasulu, Fenton & Reddy, Plant Disease 85: 208, 2001.
- Martelli, Russo, Rubino & Sabanadzovic, Archives of Virology 143: 1851, 1998.
- Miller, Damude, Robbins, Reade & Rochon, Virus Research 52: 51, 1997
- Reade, Miller, Robbins, Xiang & Rochon, Virus Research 91: 171, 2003.
- Rubino & Russo, Journal of General Virology 78: 1219, 1997.
- Rubino, Russo & Martelli, Journal of General virology, 76: 2835, 1995.
- Russo, Rubino & Martelli, Journal of Plant Pathology, 79: 125, 1997.
- Sabanadzovic, Boscia, Saldarelli, Martelli, Lafortezza & Koenig, European Journal of Plant Pathology 101: 171, 1995.