Details of DPV and References
DPV NO: 412 December 2005
Family: Bunyaviridae
Genus: Tospovirus
Species: Tomato spotted wilt virus | Acronym: TSWV
This is a revised version of DPV 39
Tomato spotted wilt virus
R. Kormelink Laboratory of Virology, Department of Plant Sciences, Wageningen University, Building 504, Binnenhaven 11, 6709 PD, Wageningen, The Netherlands
Contents
- Introduction
- Main Diseases
- Geographical Distribution
- Host Range and Symptomatology
- Strains
- Transmission by Vectors
- Transmission through Seed
- Transmission by Grafting
- Transmission by Dodder
- Serology
- Nucleic Acid Hybridization
- Relationships
- Stability in Sap
- Purification
- Properties of Particles
- Particle Structure
- Particle Composition
- Properties of Infective Nucleic Acid
- Molecular Structure
- Genome Properties
- Satellite
- Relations with Cells and Tissues
- Ecology and Control
- Notes
- Acknowledgements
- Figures
- References
Introduction
Tomato spotted wilt virus (TSWV) is the type species of the Tospovirus genus within the arthropod-borne family Bunyaviridae. The virus was first described by Samuel, Bald & Pittman (1930) and reviewed by Best (1968).
- Selected synonyms (from Best, 1968; Smith, 1972; Sakimura, 1962)
- River disease virus
- Kromnek virus
- Pineapple side rot virus
- Tomato bronze leaf virus
- Tomato bronzing virus
- Carcova virus
- cabeça virus
- Lycopersicon virus 3
- Lycopersicon virus 7
- Makhorka tip chlorosis virus
- blight virus
Particles of TSWV are spherical and membrane-bound with a diameter of approximately 80-120 nm. The virus possesses a tripartite ssRNA genome of which one segment is of negative polarity and the other two are ambisense. Virus particles are relatively unstable and during purification are greatly stabilized by the addition of reducing agents. In infected cells, mature virus particles are found in the lumen of the endoplasmic reticulum. The virus replicates in the cytoplasm where, with some isolates, fibrous inclusion bodies can be observed. TSWV is transmitted in nature by various thrips species (Thysanoptera: Thripidae) in a persistent manner, but can also readily be transmitted by mechanical inoculation. Recent reviews on tospoviruses are by Goldbach & Peters (1994, 1996), Mumford et al. (1996) and Adkins (2000).
Geographical Distribution
TSWV occurs worldwide because of its wide plant host range and the worldwide distribution of its main vector, the western flower thrips Frankliniella occidentalis. While the virus is widespread in tropical and subtropical climate zones, it is often prevalent in greenhouse cultivations in more temperate zones.
Host Range and Symptomatology
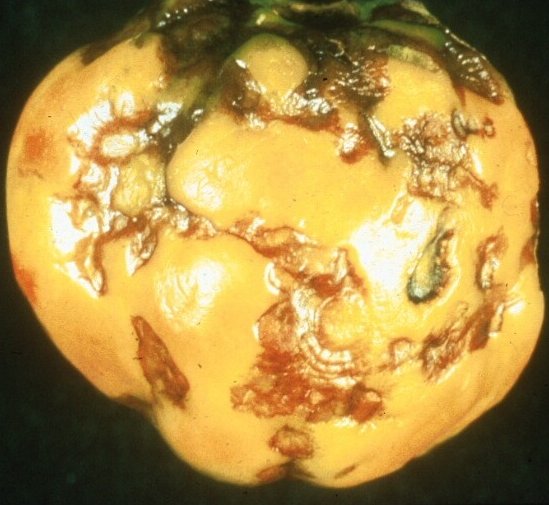
More than 800 different plant species, from 82 botanical families, including both monocotyledonous and dicotyledonous plants, have been reported to be susceptible to TSWV (Peters, http://www.dpw.wageningen-ur.nl/viro/research/hostlist.html). The virus causes significant yield losses in a large number of economically important crops e.g. groundnut, lettuce, papaya, pea, potato, sweet pepper, tobacco, tomato and in many ornamental crops, including alstroemeria, begonia, chrysanthemum, cyclamen, dahlia, gerbera, gloxinia and impatiens. Disease symptoms range from chlorosis, mottling, stunting and wilting to severe necrosis of leaf, stem and fruit tissues (Fig. 1). However, symptoms may vary within a host species according to the condition and age of the plant itself, as well as environmental conditions.
- Diagnostic species
- Petunia hybrida cvs Pink Beauty and Minstrel. Local necrotic lesions 2-4 days after inoculation; not systemic.
- Nicotiana tabacum cv Samsun NN, N. clevelandii and N. glutinosa. Local necrotic lesions, followed by systemic necrotic patterns and leaf deformation.
- Cucumis sativus. Cotyledons develop local chlorotic spots with necrotic centres, 4-5 days after inoculation.
- Vinca rosea. Local black spots, 10-14 days after inoculation, leaves sometimes with leaf yellowing and abscission; systemic mosaic and deformation.
- Tropaeolum majus. Inoculated leaves symptomless; after 8-12 days a systemic mosaic pattern of yellow and dark green specks develops, sometimes also with necrotic spots.
- Propagation species
- Tropaeolum majus and Gomphrena globosa are suitable plants for maintaining cultures. Leaves with symptoms contain much virus and are good inoculum sources. Good sources of virus for purification are the systemically infected leaves of Nicotiana rustica and N. glutinosa. Emilia sonchifolia is a good host to maintain TSWV and avoid the generation of DI-RNA during sequential mechanical inoculations (Inoue-Nagata et al., 1997).
- Assay species
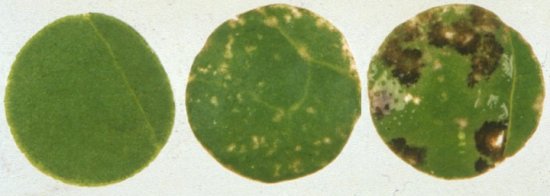
- Petunia hybrida (cvs. Pink Beauty and Minstrel) is the best local lesion host. Local necrotic lesions appear 2-4 days after inoculating in the greenhouse, but also in detached leaves in petri dishes under artificial illumination (Fig. 2).
Strains
Several strains of TSWV have been characterised and studied. During the last decade, one of the best studied ones is BR-01, originally isolated from tomato at CNPH (Centro Nacional de Pesquisa Hortaliças, Brasilia, Brazil; De Ávila et al.,1990). From this strain the complete genomic sequence has been elucidated and its cytopathology, molecular biology and vector transmission studied in detail. Most of the data used for this virus description has been obtained from studies on this strain.
Transmission by Vectors
Under natural conditions, TSWV is spread by thrips. Eight thrips species (Thysanoptera: Thripidae) have been recorded as vectors of tospoviruses, i.e. Frankliniella occidentalis Pergande (Western flower thrips), F. fusca Hinds (tobacco thrips), F. intonsa Trybom (flower thrips), F. schultzei Trybom (cotton bud thrips), F. bispinosa, Thrips palmi Karny (melon thrips), Thrips tabaci Lindeman (onion thrips) and T. setosus Moulton (De Angelis et al., 1994; Fujisawa et al., 1988; Gardner et al., 1935; Kobatake et al., 1984; Palmer et al., 1990; Pittman, 1927; Sakimura, 1963; Samuel et al., 1930; Umeya et al., 1988; Vijayalakshmi, 1994; Webb et al., 1998; Wijkamp et al., 1995; Wijkamp & Peters, 1993; Yeh et al., 1992). TSWV is transmitted by most, if not all of these species. Since the 1980s, a rapid spread of F. occidentalis has contributed to a worldwide resurgence of TSWV. Factors that have contributed to this are the concealed way of life and short life cycle of this thrips species, its ability to colonize many weed and cultivated plant species, its increased tolerance to insecticides and the global trading of thrips-infested plant material.
Transmission occurs in a propagative manner (Wijkamp et al., 1993; Ullman et al., 1993). The virus is only acquired by the first (L1) and second (L2) larval stages and can readily be transmitted in the L2 and adult stages (Van de Wetering et al., 1996; Wijkamp & Peters, 1993). For F. occidentalis, the acquisition access period (AAP50) and inoculation access periods (IAP50) needed for 50% of the thrips to acquire and inoculate TSWV are 67 min and 59 min respectively. The median latent period (LP50) decreases with increasing temperature, and ranges between 80 and 170 h (Wijkamp & Peters, 1993; Wijkamp, 1995). Once the virus is acquired, it is passed transstadially and thrips remain infectious for life.
The viral envelope proteins G1 (in more recent literature also referred to as Gc, according to the carboxy-terminal location within the precursor protein) and G2 (Gn; amino terminal location) are essential for acquisition and transmission by the vector thrips. During acquisition, G2 binds to the larval thrips gut. Furthermore, TSWV acquisition is inhibited by G2, suggesting that this protein may act as the viral ligand that mediates the attachment to receptors in the midgut epithelium of the vector thrips (Whitfield et al., 2004).
Transmission through Seed
Although the virus has been reported to be transmitted by seed on a very few occasions (Jones, 1944; Crowley, 1957), hardly any additional evidence for this has been reported. It is generally considered to be either rare or highly unlikely to occur.
Transmission by Dodder
No reports.
Serology
TSWV ribo-nucleo-proteins (RNPs) are a very good source of immunogen and good quality antisera are obtained in rabbits after three intramuscular injections of RNP preparations, scheduled for two week intervals. For all three injections RNP preparations are emulsified in Freund's incomplete adjuvant. RNPs purified by CsSO4 gradients and dialyzed prior to use for immunisation generally give the best antisera (De Ávila et al. 1990, 1993). These antisera can be used for ELISA and western blotting, as well as in situ localisation studies, with generally specific reactions and low backgrounds. Virus particles are significantly poorer immunogens, with antisera developed against purified virus preparations generally giving higher background reactions in ELISA, as well as on western blots of infected leaf samples (Feldhoff et al., 1995).
Relationships
TSWV is the type species of the genus Tospovirus in the family Bunyaviridae (Elliott, 1996). All members of this genus are distinguished on the basis of N protein serology, N protein RNA sequences and vector specificity (De Ávila et al., 1990; De Ávila et al., 1993; Goldbach & Kuo, 1996). Viruses only become recognised as a new tospovirus species in cases where the N protein sequence shows less than 90% homology to other established tospovirus species (Goldbach & Kuo, 1996). Based on these criteria 16 species have been identified to date: TSWV (De Ávila et al., 1990; De Ávila et al., 1993; De Haan et al., 1990), Tomato chlorotic spot virus (TCSV; De Ávila et al., 1990, 1993), Groundnut ringspot virus (GRSV; De Ávila et al., 1990, 1993), Impatiens necrotic spot virus (INSV; De Ávila et al., 1992; Law & Moyer, 1990; Law et al., 1991; De Haan et al., 1992a), Groundnut bud necrosis virus (GBNV; Satyanarayana et al., 1996; Reddy et al., 1992), Watermelon silver mottle virus (WSMoV; Heinze et al., 1995; Yeh & Chang, 1995), Watermelon bud necrosis virus (WBNV; Jain et al., 1998), Iris yellow spot virus (IYSV; Cortes et al., 1998; Pozzer et al., 1999), Melon yellow spot virus (MYSV; of which Physalis severe mottle virus (PhySMV) is an isolate; Kato et al., 2000; Cortez et al., 2001), Groundnut yellow spot virus (GYSV; Satyanarayana et al., 1998; Reddy et al., 1990), Groundnut chlorotic fanspot virus (GCFSV; Chen & Chiu, 1996; Chu et al., 2001), Chrysanthemum stem necrosis virus (CSNV; Bezerra et al., 1999), Zucchini lethal chlorosis virus (ZLCV; Bezerra et al., 1999), Capsicum chlorosis virus (CaCV and the synonymous Gloxinia tospovirus HT-1 and Thailand tomato tospovirus; McMichael et al., 2002; Hsu et al., 2000; Genbank Accession AF134400) and more recently Calla Lily Chlorotic Spot virus (CCSV; Chen et al., 2005) and Tomato yellow ring virus (TYRV also reported by S. Winter et al. as Tomato yellow fruit ring virus and Tomato varamin virus; Hassani-Mehraban et al., 2005; Winter et al., 2005; Ghotbi et al., 2005). According to the latest ICTV recommendations (Fauquet et al., 2005), 8 of these are presented as established species (TSWV, GRSV, TCSV, INSV, GBNV, GYSV, WSMoV and ZLCV) and 6 as tentative (IYSV, CSNV, WBNV, GCFSV, CaCV and PhySMV syn. MYSV). Several of the aforementioned species have initially been classified into four distinct serogroups, with the aid of polyclonal and monoclonal antisera directed to the viral nucleoproteins (Cortes et al., 1998; De Ávila et al., 1990; Goldbach & Kuo, 1996; Law & Moyer, 1990). Serogroup I is represented by TSWV, serogroup II by TCSV and GRSV, serogroup III by INSV and IV by GBNV, WSMV and WBNV. Members within one serogroup show serological cross-reactivity, but with the exception of members of serogroup I and II, no cross-reactivity exists between members from different serogroups. Multiple alignment of the N protein amino acid sequences confirmed the interrelationships among them and shows a clustering into American (TSWV, TCSV, GRSV, INSV, CSNV, ZLCV) and EuroAsian (GBNV, WSMoV, WBNV, IYSV, MYSV) branches. GYSV and GCFSV show only a distant relationship to the EuroAsian branch. On a higher taxonomic level, there is no serological relationship between tospoviruses and members from other genera of the family Bunyaviridae. However, a low but significant level of amino acid sequence homology was found for the precursor to the glycoproteins and for the RNA polymerase, between TSWV and members of the OrthoBunyavirus genus (Cortez et al., 2002; De Haan et al., 1991; Kormelink et al., 1992a).
Stability in Sap
Virus particles are unstable, with a thermal inactivation point (10 min) of 40-46°C, a longevity in vitro at room temperature of 2-5 h and a dilution end-point between 2x10-2 and 1x10-4. During purification virus particles are greatly stabilized by the addition of reducing agents such as 0.01 M Na2SO3. For TSWV, yields are usually about 100-1000 mg per 100 g leaf material of Nicotiana rustica.
Purification
Complete virus particles are isolated at 4°C from systemically infected N. rustica leaves, essentially as previously published (Gonsalves & Trujillo, 1986; Tas et al. ,1977). Harvested leaves are ground in 3 ml of extraction buffer (0.01 M sodium sulphite; 0.1 M sodium phosphate pH 7.0) per g leaf material in a blender by giving five to ten short pulses at medium speed. The homogenate is filtered through cheesecloth and the extract is centrifuged at 10,000 g for 15 min. The pellet obtained is gently homogenized for 30 min in 1 ml resuspension buffer (0.01 M sodium sulphite; 0.01 M sodium phosphate pH 7.0) per g initial leaf material using a small pestle and a magnetic stirrer. The suspension is clarified by centrifugation at 8,000 g for 15 min and the supernatant is collected and centrifuged at 100,000 g for 30 min. The resulting pellet is homogenized in 5 ml resuspension buffer per 100 g of initial leaf material and 2.5 ml is layered per 10-40% sucrose gradient. After centrifugation for 45 min at 70,000 g in a Beckman SW28 rotor, the opalescent zone containing virus particles is collected, diluted 1:1 with resuspension buffer and concentrated by centrifuging for 1 h at 100,000 g. The pellet, containing virus particles, is resuspended in sterile double-distilled water at a concentration of approximately 10 mg/ml. Virus from these preparations support replication and transcription in vitro (Van Knippenberg et al., 2002). In some experiments the sucrose gradient at the end of the procedure can be omitted, and the preparation is homogenized in sterile double-distilled water. This also yields infectious virus suitable for inoculation of protoplasts (Kikkert et al., 1997). Virus preparations are kept on ice and used immediately, or promptly frozen in liquid nitrogen and stored at -80°C. Before use, samples are thawed slowly on ice.
Properties of Particles
TSWV particles sediment at 530-583 S (s20,w; Best, 1968; Black et al., 1963).
Particle Structure
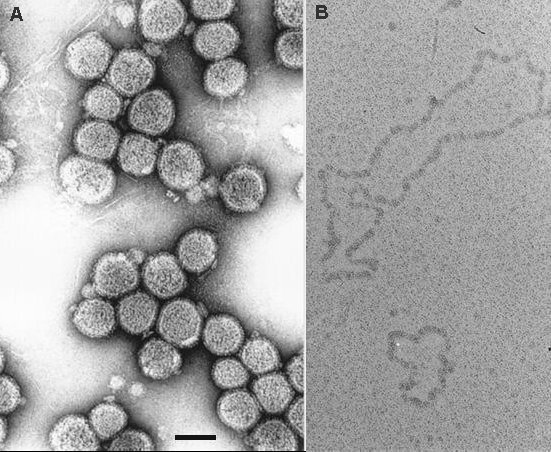
TSWV have particles that are spherical and membrane bound (80-120 nm; Fig. 3a and Fig. 4). The particles are covered with surface projections of about 5-10 nm in length, that consist of two glycoproteins, G1 (78 KDa) and G2 (58 KDa) (Mohamed et al., 1973; Tas et al., 1977). An additional smaller version of the G2 protein of about 52 KDa is often observed in purified TSWV preparations. In purified TSWV from infected plants, G2 exists as monomers, homodimers and heterodimers with G1 (Whitfield et al., 2004). The core consists of pseudo-circular RNPs (Fig. 3b), each consisting of a viral RNA segment tightly wrapped in molecules of the nucleoprotein (N, 29 KDa) and minor amounts of a large protein (L, 331.5 KDa), the putative viral RNA polymerase (Mohamed, 1981; Tas et al., 1977; Van den Hurk et al., 1977; Van Poelwijk et al., 1993; Verkleij & Peters, 1983). For the N protein, reported sizes range between 28.7 and 30.7 KDa. Purified particles, as well as RNPs, but not the RNA, are infective when inoculated onto plants. Only enveloped particles are transmitted by thrips (Nagata et al., 2000; Resende et al., 1991; Wijkamp, 1995).
Particle Composition
Nucleic acid: TSWV contains a tripartite RNA genome with a total size of 16.6 kb and a G+C content of 34.4%. The RNA segments are linear and single stranded, of which one element is of negative polarity and two are ambisense. The largest (L) RNA segment has 8897 nucleotides (nt), the medium (M) sized segment has 4821 nt, the small (S) RNA segment has 2918 nt and together contain 5 transcription units (De Haan et al., 1989, 1990, 1991; Kormelink et al., 1992a). Viral (v) and viral complementary (vc) strands of all three genomic RNA segments are found in virus preparations; the v strands are in excess over vc strands, possibly reflecting the difference in amounts synthesized in infected plants (Kormelink et al., 1992b).
Protein: The virus consists of three major structural proteins: the nucleoprotein with a mol. wt of 29 kDa and two glycoproteins, G1 and G2, of 78 and 58 kDa respectively; as estimated from polyacrylamide gel electrophoresis and confirmed by deduction from the amino acid sequence of the encoding transcription units. Particles contain about 65% protein, 20% lipid, 7% carbohydrate and 5% RNA (Best, 1968).
Genome Properties
The complete nucleotide sequence of the tripartite genome of TSWV has been determined (De Haan et al., 1989, 1990, 1991; Kormelink et al., 1992a). A schematic representation of the genome organisation of TSWV is shown in Fig. 5. This organisation applies to all members of the Tospovirus genus studied so far. The large (L) RNA segment contains 8897 nucleotides (nt) and is of negative polarity. The viral complementary (vc) strand contains one large ORF, encoding a protein of 331.5 KDa. Based on sequence homology with the Bunyamwera virus L protein and influenza virus PB1 protein, alongside in vitro TSWV RNA synthesis inhibition by antibodies directed against the L protein, this protein is thought to represent the viral RNA-dependent RNA polymerase (De Haan et al., 1990; Chapman et al., 2003). The medium (M) RNA segment contains 4821 nt and has an ambisense gene arrangement (Kormelink et al., 1992a). This RNA segment encodes, in the viral (v) sense, a nonstructural protein (NSm) of 33.6 KDa, which is implicated in cell-to-cell movement (Kormelink et al., 1994; Storms et al., 1995, 1998), and in the vc sense, a 127.4 KDa protein which is the precursor to the glycoproteins (G1 and G2). The small (S) RNA segment contains 2916 nt and, like the M RNA, has an ambisense gene arrangement. This genome segment encodes a nonstructural (NSs) protein of 52.4 KDa in the v sense, which acts as a suppressor of silencing (Takeda et al., 2002; Bucher et al., 2003), and the nucleoprotein (N) of 28.8 KDa in the vc sense (De Haan et al., 1990; Kormelink et al., 1991). All viral genes become expressed by the synthesis of mRNA molecules. No subgenomic RNA molecules derived from L RNA have been detected, suggesting that the mRNA for the L ORF is of almost full length size. The M and S RNA segments each give rise to two subgenomic RNA molecules, transcribed from complementary strands, reflecting the ambisense gene arrangement of both segments (Kormelink et al., 1992a, 1992b; De Haan et al., 1990). Analysis of the RNA contents of the virus particles and nucleocapsid preparations has shown that these subgenomic mRNA molecules are not encapsidated into RNPs and, as a result, are not present in virus particles. Primer extension and nucleotide sequence analyses of the subgenomic N, NSs and NSm mRNAs revealed the presence of heterogeneous non-viral sequences, 12-20 nt in length, at the 5' ends of the mRNA molecules (Duijsings et al. 1999, 2001; Kormelink et al., 1992c; Van Knippenberg et al., 2002; Van Poelwijk et al., 1996), indicating that in TSWV “cap-snatching” (Braam et al., 1983; Plotch et al., 1981; Ulmanen et al., 1981) is the mechanism by which transcription of the viral genome is initiated. It has not been shown whether the endonuclease activity involved in the cap-snatching is encompassed in the viral polymerase (L protein).
About 65-70 nt of the L, M and S RNA molecules are complementary at the 5' and 3' ends, and this results in the formation of stable “panhandle” structures, causing the RNA segments to appear as pseudo-circular structures (Fig.3b). The 5'- and 3'-terminal sequences of all three RNA segments are identical for the first eight nucleotides, a typical feature of negative-stranded, segmented RNA viruses (e.g. Orthomyxoviridae, Bunyaviridae, Arenaviridae and the tenuiviruses); this is thought to have an important function in genome replication and transcription (De Haan et al., 1989). The non-coding intergenic regions of both the M and S RNA segments contain an internal, inverted sequence of A- and U-rich stretches that can be folded into a hairpin structure. The 3' ends of the S RNA-derived N and NSs mRNAs have been shown to overlap at their 3' ends and contain the predicted hairpin structure (Van Knippenberg et al., 2005). Furthermore, a conserved sequence (CAAACUUUGG) has been found at the top of the hairpin of the TSWV M and S RNA segments. Both structural features have been suggested a possible function in termination of transcription.
- DI RNA
- RNA molecules of subgenome length, derived from the L RNA, have been found repeatedly as a result of serial mechanical passages of TSWV at high inoculum concentration and low temperature regimes (Inoue-Nagata et al., 1997; Resende et al., 1991). These defective RNA molecules have been shown to interfere with the replication of the wild type genome (Resende et al., 1991), and hence cause attenuation of symptom expression. All DI RNA molecules studied have been shown to retain the genomic 5' and 3' ends, suggesting that these sequences are essential for replication, encapsidation and subsequent packaging into virus particles (Inoue-Nagata et al., 1998; Resende et al., 1992). The mode by which the internal deletions are generated is unknown. Transmission of TSWV by the thrips vector is negatively affected by the additional presence of DI RNAs (Wijkamp, 1995; Nagata et al., 2000).
Relations with Cells and Tissues
TSWV is found in almost all tissues and organs following systemic infection of plants (Ie, 1973; Francki & Grivell, 1970; Kitajima, 1965; Kitajima et al., 1992). Mature virus particles mainly accumulate in clusters in the cisternae of the rough endoplasmic reticulum (RER) and consist of spherical lipid-bound particles, 80-120 nm in diameter, covered with spike projections (Fig. 6; Best & Palk, 1964; Best & Katekar, 1964; Francki & Grivell, 1970; Francki & Hatta, 1981; Francki et al., 1984; Ie, 1964; Kikkert et al., 1999; Kitajima, 1965; Martin, 1964; Van Kammen et al., 1966).
In addition to mature virus particles, specific cytopathic structures are associated with TSWV infection. One type is characterised by dark diffuse amorphous masses (Fig. 7), sometimes called viroplasm, with locally electron-dense striated spots and found freely dispersed in the cytoplasm (Francki and Grivell, 1970; Francki and Hatta, 1981; Francki et al., 1984; Ie, 1971; Kitajima, 1965; Kitajima et al., 1992; Milne, 1970). These dense spots have a diameter slightly smaller than that of mature virus particles (Ie, 1971) and are proteinaceous in nature; they are suggested to consist of ribonucleoprotein and to be a normal developmental stage in the formation of TSWV particles (Milne, 1970; Ie, 1971). This idea receives support in studies of morphologically defective TSWV isolates (Ie, 1982, Resende et al., 1991; Verkleij & Peters, 1983), in which purified infectious fractions of TSWV were found to resemble the amorphous masses in infected plant cells. It was therefore suggested that these masses consist of aggregates of nucleocapsids that are not enveloped, either because they are at an intermediate stage of development or because they represent morphologically defective TSWV that for some reason cannot produce enveloped particles (Ie, 1982; Resende et al., 1991; Verkleij & Peters, 1983). The second type of cytopathic structure associated with tospovirus infections are fibrous (Fig. 8; Francki and Grivell, 1970; Francki et al., 1984; Kitajima et al., 1992; Kormelink et al., 1991) and in the case of TSWV consist of elongated flexible filaments. These structures have been shown to contain one of the nonstructural proteins of TSWV, the NSs protein (Kitajima et al., 1992; Kormelink et al., 1991).
During an early stage of infection, doubly-enveloped particles are found in the cytoplasm (Francki et al., 1984; Kikkert et al., 1999; Kitajima et al., 1992; Milne, 1970). These are the result of nucleocapsids that have become enwrapped by Golgi cisternae, appearing as paired parallel membranes, to form doubly enveloped particles (DEVs; Fig. 9; Kikkert et al., 1999). Subsequent fusion of DEVs with each other and with ER membranes finally lead to the formation of a cluster of singly enveloped particles (SEVs) in the cisternae of RER (Kikkert et al., 1999). Virus particles have not been observed in the lumen of the Golgi complex or vacuoles.
During early stages of the infection, moreover, tubular structures have been observed extending from plasmodesmata into newly infected cells. These structures have been shown to be composed of another nonstructural protein, NSm (Kormelink et al., 1994; Storms et al., 1995). Microinjection of fluorescent dyes into parenchyma cells of transgenic plants expressing this protein, has shown that the size exclusion limit of plasmodesmata was modified (Storms et al., 1998). Hence, the NSm protein has been implicated in the cell-to-cell movement of non-enveloped infectious ribonucleocapsid structures of TSWV.
In viruliferous individuals of the thrips F. occidentalis, large amounts of the N and NSs proteins are found in the salivary glands. Additionally, both proteins are present in midgut epithelium and in muscle cells associated with the midgut epithelium (Tsuda et al., 1996; Nagata et al., 1999; Ohnishi et al., 2001). The ligaments between the midgut and the salivary glands appears to be a route of the virus to invade the salivary glands (De Assis Filho et al., 2002; Kritzman et al., 2002; Nagata et al., 1999, 2002). Mature virus particles are observed in vesicles in the salivary glands and in massive numbers in the salivary gland ducts; suggesting that the salivary glands are the major site of replication (Ullman et al., 1992 and 1993; Wijkamp et al.,1993). In adults that have fed on infected leaf material, virus is never found beyond midgut epithelial cells (Ohnishi et al., 2001; Ullman et al., 1992).
Ecology and Control
Weeds play an important role in the spread and survival of TSWV; forming a virus reservoir from where viruliferous thrips can migrate to crop fields that there after become heavily infected (Bond et al., 1983; Cho et al., 1986; Kobatake et al., 1984). Since thrips have become resistant against most of the insecticides used they have become hard to control (Brødsgaard, 1994; Zhao et al., 1995). Therefore, nowadays management of TSWV infections in the field is based on a combination of thrips management, weed control, use of resistant crop varieties and other measurements. So far, only a limited number of resistant genes are available for introduction into plant breeding programs; SW5 is one of them (Boiteux et al., 1993; Boiteux and Gordiano, 1993; Stevens, 1994). This gene has been isolated, characterised and after transformation to non-resistant hosts of TSWV, demonstrated to confer resistance to TSWV (Brommonschenkel et al., 1997, 2000; Folkertsma et al., 1999; Spassova et al., 2001). Next to the use of natural resistance genes, transgenic resistance strategies have been developed and proven successful either by the transformation of hosts with coding or non-coding sequences of the N and NSm genes (De Haan et al., 1992b; Prins et al., 1995, 1997) or by transformation with so-called 'plantibodies' specific against the viral N protein (Prins et al., 2000, 2005) or with so-called 'aptamers', i.e. dominant interfering peptides (Rudolph et al., 2003). The latter two approaches have both shown to confer resistance to a wider range of tospoviruses.
Figures

Schematic representation of a TSWV particle. N, nucleoprotein; L, viral RdRp; G1 and G2 represent the viral spike proteins.

Genomic organisation of the tripartite RNA genome of TSWV. Circles at the 5'-end of TSWV mRNAs indicate non-viral sequences used to initiate transcription of the viral mRNAs. Bars represent open reading frames. vRNA, viral RNA; vcRNA, viral complementary RNA.

Cytopathology of TSWV-infected leaf tissue (from Kormelink, 1995): clustered TSWV virions (V) accumulating in large vesicles. Bar represents 100 nm.

Cytopathology of TSWV-infected leaf tissue (from Kormelink, 1995): electron dense masses (DM) consisting of non-enveloped nucleocapsid aggregates in the cytoplasm;. Bar represents 100 nm.
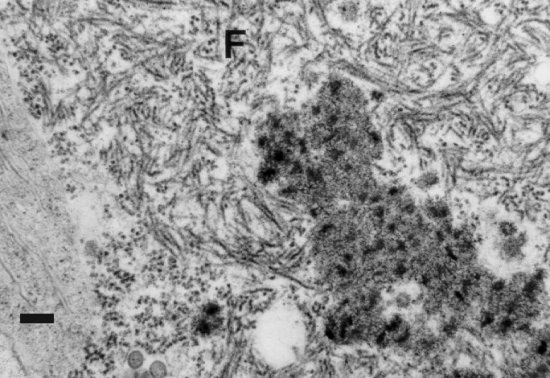
Cytopathology of TSWV-infected leaf tissue (from Kormelink, 1995): fibrous structures (F) in the cytoplasm of a TSWV-infected cell. Bar represents 100 nm.

Cytopathology of TSWV-infected leaf tissue (from Kormelink, 1995): virus particles bound by two membranes (doubly enveloped virus particles, DEVs). Bar represents 100 nm.
References list for DPV: Tomato spotted wilt virus (412)
- Adkins, Molecular Plant Pathology 1: 151, 2000.
- Best, Advances in Virus Research 13: 65, 1968.
- Best & Katekar, Nature 203: 671, 1964.
- Best & Palk, Virology 23: 445, 1964.
- Bezerra, Resende, Pozzer, Nagata, Kormelink & De Ávila, Phytopathology, 89: 823, 1999.
- Black, Brakke & Vatter, Virology 20: 120,1963.
- Boiteux, Nagata, Dutra & Fonseca, Euphytica 67: 89, 1993.
- Boiteux & Giordano, Euphytica 71: 151, 1993.
- Bond, Whitman & Black, Phytopathology 73: 499, 1983.
- Braam, Ulmanen & Krug, Cell 34: 609, 1983.
- Brødsgaard, Journal of Economic Entomology 87: 1141, 1994.
- Brommonschenkel, Frary, Frary & Tanksley, Molecular Plant Microbe Interactions 13: 1130, 2000.
- Brommonschenkel & Tanksley, Molecular and General. Genetics 256: 212, 1997.
- Bucher, Sijen, De Haan, Goldbach & Prins, Journal of Virology 77: 1329, 2003.
- Chapman, Hilson & German, Intervirology 46: 177, 2003.
- Chen, Chen, Lin, Yeh & Hsu, Plant Disease 89: 440, 2005.
- Chen & Chiu, Acta Horticulturae 432: 57, 1996.
- Cho, Mau, Gonsalves & Mitchell, Plant Disease 70: 1014, 1986.
- Chu, Chao, Peng, Lin, Chen & Yeh, Phytopathology 91: 856, 2001.
- Cortes, Livieratos, Derks, Peters & Kormelink, Phytopathology 88:1276, 1998.
- Cortez, Saaijer, Wongjkaew, Pereira, Goldbach, Peters & Kormelink, Archives of Virology 146: 265, 2001.
- Cortez, Aires, Pereira, Goldbach, Peters & Kormelink, Archives of Virology 147: 2313, 2002.
- Crowley, Australian Journal of Biological Science 10: 449, 1957.
- De Angelis, Sether & Rossignol, Environmental Entomology. 22: 1308, 1994.
- De Assis Filho, Naidu, Deom & Sherwood, Phytopathology 92: 729, 2002.
- De Ávila, Huguenot, Resende, Kitajima, Goldbach & Peters, Journal of General Virology 71: 2801, 1990.
- De Ávila, De Haan, Kitajima, Kormelink, Resende, Goldbach & Peters, Journal of Phytopathology 134: 133, 1992.
- De Ávila, De Haan, Kormelink, Resende, Goldbach & Peters, Journal of General Virology 74:153, 1993.
- De Haan, Wagemakers, Peters & Goldbach, Journal of General Virology 70: 3469, 1989.
- De Haan, Wagemakers, Peters & Goldbach, Journal of General Virology 71: 1001, 1990.
- De Haan, Kormelink, Resende, Van Poelwijk, Peters & Goldbach, Journal of General Virology 72: 2207, 1991.
- De Haan, De Ávila, Kormelink, Westerbroek, Gielen, Peters & Goldbach, FEBS Letters 306: 27, 1992a.
- De Haan, Gielen, Prins, Wijkamp, Van Schepen, Peters, Van Grinsven & Goldbach, Bio/Technology 10: 1133, 1992b.
- Duijsings, Kormelink & Goldbach, Journal of Virology 73: 5172, 1999.
- Duijsings, Kormelink & Goldbach, EMBO Journal 20: 2545, 2001.
- Elliott (ed.), "The Bunyaviridae", New York: Plenum Press, 1996.
- Fauquet, Mayo, Maniloff, Desselberger & Ball (eds), Eight Report of the International Committee on the Taxonomy of Viruses, London: Elsevier Academic Press, 2005.
- Feldhoff, Kikkert, Kormelink, Krczal, Goldbach & Peters, Archives of Virology 142: 781, 1995.
- Folkertsma, Spassova, Prins, Stevens, Hille & Goldbach, Molecular Breeding 5: 197, 1999.
- Francki & Grivell, Virology 42: 969, 1970.
- Francki & Hatta, in Plant Virus Infections and Comparative Diagnosis, p.491, ed. Kurstak, Amsterdam: Elsevier/North-Holland, 1981.
- Francki, Milne & Hatta, in Atlas of Plant Viruses, Vol.1, p.101, Boca Raton: CRC Press, 1984.
- Fujisawa, Tanaka & Ishii, Annals of the Phytopathological Society of Japan 54: 392, 1988.
- Gardner, Tompkins & Whipple, Phytopathology 25: 17, 1935.
- Ghotbi, Shahraeen & Winter, Plant Disease 89: 425, 2005.
- Goldbach & Kuo, Acta Horticulturae 432: 21, 1996.
- Goldbach & Peters, Seminars in Virology 5: 113, 1994.
- Goldbach & Peters, in The Bunyaviridae, p.129, ed. Elliott, New York, Plenum Press,1996.
- Gonsalves & Trujillo, Plant Disease 70: 501, 1986.
- Harrison & Robinson, in Handbook of Plant Virus Infections and Comparative Diagnosis, p.515, ed. Kurstak, Amsterdam: Elsevier/North-Holland, 1981.
- Hassani-Mehraban, Saaijer, Peters, Goldbach & Kormelink, Phytopathology 95: 852, 2005.
- Heinze, Maiss, Adam & Casper, Phytopathology 85: 683, 1995.
- Hsu, Ueng, Chu, Ye & Yeh, Journal of General Plant Pathology 66: 167, 2000.
- Ie, Netherlands Journal of Plant Pathology 70: 114, 1964.
- Ie, Virology 2: 468, 1971.
- Ie, Netherlands Journal of Plant Pathology 79: 387, 1973.
- Ie, Journal of General Virology 59: 387, 1982.
- Inoue-Nagata, Kormelink, Nagata, Kitajima, Goldbach & Peters, Phytopathology 87: 1168, 1997.
- Inoue Nagata, Kormelink, Sgro, Nagata, Kitajima, Goldbach & Peters, Virology 248: 342, 1998
- Jain, Pappu, Pappu, Reddy & Vani, Archives of Virology 143: 1637, 1998.
- Jones, Phytopathology 34: 941, 1944.
- Kato, Hanada & Kameya-Iwaki, Phytopathology 90: 422, 2000.
- Kikkert, Van Poelwijk, Storms, Bloksma, Karsies, Kormelink & Goldbach, Journal of General Virology 78: 1755, 1997.
- Kikkert, Van Lent, Storms, Bodegom, Kormelink & Goldbach, Journal of Virology 73; 2288, 1999.
- Kitajima, Virology 26: 89, 1965.
- Kitajima, De Ávila, Resende, Goldbach & Peters, Journal of Submicroscopic Cytology and Pathology 24: 1, 1992.
- Kobatake, Osaki & Inouye, Annals of the Phytopathological Society of Japan 50: 541, 1984.
- Kormelink, PhD Thesis, Wageningen, 1995.
- Kormelink, Kitajima, De Haan, Zuidema, Peters & Goldbach, Virology 181: 459, 1991.
- Kormelink, De Haan, Meurs, Peter & Goldbach, Journal of General Virology 73: 2795, 1992a.
- Kormelink, De Haan, Peters & Goldbach, Journal of General Virology 73: 687, 1992b.
- Kormelink, Van Poelwijk, Peters & Goldbach, Journal of General Virology 73: 2125, 1992c.
- Kormelink, Storms, Van Lent, Peters & Goldbach, Virology 200: 56, 1994.
- Kritzman, Gera, Raccah, Van Lent & Peters, Archives of Virology 147: 2143, 2002.
- Law & Moyer, Journal of General Virology 71: 933, 1990.
- Law, Speck & Moyer, Journal of General Virology 72: 2597, 1991.
- Martin, Virology 22: 645, 1964.
- McMichael, Perley & Thomas, Australasian Plant Pathology 31: 231, 2002.
- Milne, Journal of General Virology 6: 267, 1970.
- Mohamed, Journal of General Virology 53: 197, 1981.
- Mohamed, Randles & Francki, Virology 56: 12, 1973.
- Mumford, Barker & Wood, Annals of Applied Biology 128: 159, 1996.
- Nagata, Inoue-Nagata, Prins, Goldbach & Peters, Phytopathology 90: 454, 2000.
- Nagata, Inoue-Nagata, Smid, Goldbach & Peters, Journal of General Virology 80: 507, 1999.
- Nagata, Inoue-Nagata, Van Lent, Goldbach & Peters, Journal of General Virology 83: 663, 2002.
- Ohnishi, Knight, Hosokawa, Fujisawa & Tsuda, Phytopathology 91: 1149, 2001.
- Palmer, Reddy, Wightman & Ranga Rao, International Arachis Newsletter 7: 24, 1990.
- Pittman, Council of Science of India Research Bulletin 1: 74, 1927.
- Plotch, Bouloy, Ulmanen & Krug, Cell 23: 847, 1981.
- Pozzer, Bezerra, Kormelink, Prins, Peters, Resende & De Avila, Plant Disease 83: 345, 1999.
- Prins, De Haan, Luyten, Van Veller, Van Grinsven & Goldbach, Molecular Plant Microbe Interactions 8: 85, 1995.
- Prins, Kikkert, Ismayadi, De Grauw, De Haan & Goldbach, Plant Molecular Biology 33: 235, 1997.
- Prins, Lohuis, Schots & Goldbach, Journal of General Virology 86: 2107, 2005.
- Prins, Schouten, Schots & Goldbach, in Molecular Farming p.207, eds. Toutant & Balázs Paris: INRA editions, 2000.
- Reddy, Sudarshana, Ratna, Reddy, Amin, Kumar & Murphy, Proceedings of the USDA Workshop, Beltsville: 77, 1990.
- Reddy, Ratna, Sudarshana, Poul & Kumar, Annals of Applied Biology 120: 279, 1992.
- Resende, De Haan, De Ávila, Kitajima, Kormelink, Goldbach & Peters, Journal of General Virology 72: 2375, 1991.
- Resende, De Haan, Van der Vossen, De Ávila, Goldbach & Peters, Journal of General Virology 10: 2509, 1992.
- Rudolph, Schreier & Uhrig, Proceedings of the National Academy of Science, USA 100: 4429, 2003.
- Sakimura, in Biological Transmission of Disease Agents p.37, ed. Maramorosch, New York: Academic Press, 1962.
- Sakimura, Phytopathology 53: 412, 1963.
- Samuel, Bald & Pittman, Council of Science of India Research Bulletin 44: 1, 1930.
- Satyanarayana, Mitchell, Reddy, Brown, Kresovich, Jarret, Naidu & Demski, Archives of Virology 141: 85, 1996.
- Satyanarayana, Gowda, Lakshminarayana Reddy, S.E. Mitchell, Dawson & Reddy, Archives of Virology 143: 353, 1998.
- Smith, in Textbook of Plant Virus Diseases 3rd edition, p. 245, ed. Smith, London: Longman, 1972.
- Spassova, Prins, Folkertsma, Klein-Lankhorst, Hille, Goldbach & Prins, Molecular Breeding 7:151, 2001.
- Stevens, Scott & Gegerlich, Euphytica 80: 79, 1994.
- Storms, Kormelink, Van Lent, Peters & Goldbach, Virology 214: 485, 1995.
- Storms, Van der Schoot, Prins, Kormelink, Van Lent & Goldbach, The Plant Journal 13: 131, 1998
- Takeda, Sugiyama, Nagano, Mori, Kaido, Mise, Tsuda & Okuno, FEBS Letters 532: 75, 2002.
- Tas, Boerjan & Peters, Journal of General Virology 36: 267, 1977.
- Taylor & Brown, Nematode Vectors of Plant Viruses, Wallingford: CAB International, 1997.
- Tsuda, Fujisawa, Ohnishi, Hosokawa & Tomaru, Phytopathology 86: 1199, 1996.
- Ullman, Cho, Mau, Westcot & Custer, Phytopathology 82: 1333, 1992.
- Ullman, German, Sherwood, Westcot, & Cantone, Phytopathology 83: 456, 1993.
- Ulmanen, Broni & Krug, Proceedings of the National Academy of Science, USA 12: 7355, 1981.
- Umeya, Kudo & Miyazaki, Pest Thrips in Japan, Tokyo: Zenkoku Noson Kyoiku Kyokai Publishing Co., 1988.
- Van de Wetering, Goldbach & Peters, Phytopathology 86: 900, 1996.
- Van den Hurk, Tas & Peters, Journal of General Virology 36: 81, 1977.
- Van Kammen, Henstra & Ie, Virology 30: 574, 1966.
- Van Knippenberg, Goldbach & Kormelink, Virology 303: 278, 2002.
- Van Knippenberg, Goldbach & Kormelink, Virus Research 110:125, 2005.
- Van Poelwijk, Boye, Oosterling, Peters & Goldbach, Virology 197: 468, 1993.
- Van Poelwijk, Kolkman & Goldbach, Archives of Virology 141: 177, 1996.
- Verkleij & Peters, Journal of General Virology 64: 677, 1983.
- Vijayalakshmi, PhD Thesis, Andhra Pradesh Agricultural University, 1994.
- Webb, Tsai & Mitchell, In Recent Progress in Tospovirus and Thrips Research p. 67, eds. Peters & Goldbach, Wageningen , The Netherlands, 1998.
- Whitfield, Ullman & German, Journal of Virology 78:13197, 2004.
- Wijkamp, PhD Thesis, Wageningen, 1995.
- Wijkamp & Peters, Phytopathology 83: 986, 1993.
- Wijkamp, Van Lent, Kormelink, Goldbach & Peters, Journal of General Virology 74: 341, 1993.
- Wijkamp, Almarza, Goldbach & Peters, Phytopathology 85: 1069, 1995.
- Winter, Shahraeen, Koerbler & Lesemann, in New Disease Reports Vol.11, 2005.
- Yeh & Chang, Phytopathology 85: 58, 1995.
- Yeh, Lin, Cheng, Jih, Chen & Chen, Plant Disease 76: 835, 1992.
- Zhao, Liu, Brown & Knowles, Journal of Economic Entomology 88: 1164, 1995.