Details of DPV and References
DPV NO: 413 June 2006
Family: Caulimoviridae
Genus: Cavemovirus
Species: Cassava vein mosaic virus | Acronym: CsVMV
Cassava vein mosaic virus
P. Marmey UMR DGPC, Résistance des Plantes, Montpellier, France
C.M. Fauquet ILTAB, Donald Danforth Plant Science Center, St. Louis, USA
Contents
- Introduction
- Main Diseases
- Geographical Distribution
- Host Range and Symptomatology
- Strains
- Transmission by Vectors
- Transmission through Seed
- Transmission by Grafting
- Transmission by Dodder
- Serology
- Nucleic Acid Hybridization
- Relationships
- Stability in Sap
- Purification
- Properties of Particles
- Particle Structure
- Particle Composition
- Properties of Infective Nucleic Acid
- Molecular Structure
- Genome Properties
- Satellite
- Relations with Cells and Tissues
- Ecology and Control
- Notes
- Acknowledgements
- Figures
- References
Introduction
Cassava vein mosaic disease was first described by Costa (1940). Cassava vein mosaic virus (CsVMV) was described by Kitajima & Costa (1966) and Lin & Kitajima (1980).
The virus has been reported from many states in Brazil and is prevalent in the northeastern states. Only Manihot esculenta is known to be affected. The virus has isometric particles 50 nm in diameter and its genome is composed of DNA. Dense inclusion bodies can be observed in the cytoplasm of infected cells. The virus is transmitted by vegetative propagation.
Main Diseases
Little is known about disease loss. Symptoms include a chlorosis of the veins, which can either appear as a chevron pattern or coalesce to form a mosaic pattern. Some leaves show a mosaic pattern over the entire leaf.
Geographical Distribution
The virus is widespread throughout the northeastern region of Brazil, although there are also reports from other regions of the country. According to Calvert et al. (1995), the semi-arid region of the northeastern states of Cereà, Pernambuco and Bahia was the region where cassava plants infected with CsVMV were most prevalent; fields could be found with 50 to 100% of the plants infected.
Host Range and Symptomatology
Cassava vein mosaic disease was first reported in Manihot esculenta by Costa (1940). So far, cassava is the only known host for CsVMV.
Expression of symptoms is variable and symptoms may fade with age or not be expressed during certain times of the year. Cassava plants infected with CsVMV display a range of symptoms including a chlorosis following the veins; the chlorosis can coalesce to form a mosaic pattern (Fig. 1 & Fig. 2). Some leaves show a mosaic pattern over the entire leaf. Leaf distortion and epinasty are often observed on young leaves.
Strains
There have been very few virus isolations and therefore there are no distinct CsVMV strains on record. The isolate first sequenced was from Brazil (Calvert et al., 1995). Cloned full-length DNA, designated pCVMV141, may be regarded as representing the type isolate.
Transmission by Vectors
There is no conclusive evidence for transmission by vectors.
Transmission through Seed
There is no known transmission of the virus through seed. CsVMV has been shown to be transmitted by vegetative propagation. The virus is easily transmitted through the stem cuttings used as reproductive material of cassava.
Serology
The virus can be detected by standard serological methods (Lin & Kitajima, 1980).
Relationships
There are no data on serological relationships with other viruses.
Stability in Sap
Leaf sap contains few virions (Lin & Kitajima, 1980).
Purification
A protocol for CsVMV particle purification from infected cassava leaves was published by Lin & Kitajima (1980). Tissue was ground in extraction buffer (0.5 M KPO4, containing 0.75% Na2SO3, 2.5% Triton X-100 and 1 M urea) before centrifugation to get rid of debris. The debris was re-extracted twice. The combined extracts were ultracentrifuged at 27,000 rpm for 90 min. to sediment the virus. The resuspended virus was centrifuged in a sucrose gradient at 24,000 rpm for 2 h. An opalescent band at 30-39 mm was observed and contained virus particles.
Properties of Particles
The sedimentation coefficient, S°20,w, was estimated to be about 246S (Lin & Kitajima, 1980).
Particle Structure
Virions are not enveloped. They are isometric, appear to be spherical, and are 45-50 nm in diameter (Fig. 3).
Particle Composition
The virus particles contain circular dsDNA (Lin & Kitajima, 1980), comprising 8158 bp (Calvert et al., 1995) or 8159 bp (de Kochko et al., 1998).
Genome Properties
The two complete sequences of CsVMV (an unnamed isolate, accession number U20341, Calvert et al., 1995; an infectious clone, accession number U59751, de Kochko et al., 1998) are slightly different regarding the position of two open reading frames (ORF). They are however similar for the first three ORFs that contain essential genes for structure and replication of the virus.
The particularity of CsVMV is the arrangement of genes in the genome, which is distinct from that of caulimoviruses and badnaviruses. It was proposed that CsVMV be the type species of the genus Cavemovirus, a new genus of the plant pararetrovirus family Caulimoviridae (Hull et al., 2005). The genomic organization of accession number U59751 (Fig. 4) was adopted by the International Committee on the Taxonomy of Viruses as being typical of the genus.
As in the genomes of other pararetroviruses, CsVMV contains a sequence that is complementary to a plant cytoplasmic tRNA. The numbering of the CsVMV genome starts at the 5'-end of this putative primer-binding site.
ORF 1 potentially encodes a protein of 1372 amino acids with a molecular mass of 186 kDa. A putative RNA-binding site domain (CXCX2CX4HX4C), found in all pararetroviruses capsid proteins, is found at the location aa 739-754. This motif is followed by a consensus sequence (aa 972-1105) similar to the intercellular transport proteins of the caulimoviruses. This arrangement of MP with respect to CP is unique.
ORF 2 overlaps ORF 1 by 14 nucleotides. The putative encoded protein of 71 amino acids, with a molecular mass of 8.8 kDa, has no homology with other proteins; its role is unknown.
The putative protein encoded by ORF 3 is 652 amino acids long with a molecular mass of 77 kDa. It contains sequences similar to protease, reverse transcriptase and RNase H consensus regions. The presence of such proteins makes CsVMV similar to other plant pararetroviruses.
Calvert et al. (1995) found two more ORFs. ORF 4 and ORF 5 overlap the preceding ORF. ORF 4 and ORF 5 encode putative proteins of 201 and 220 amino acids, with molecular masses of 24 and 26 kDa, respectively. There is no similarity with known proteins encoded by caulimoviruses or badnaviruses.
The sequence described by de Kochko et al. (1998), derived from the complete sequence of an infectious clone, differs from the one described by Calvert et al. (1995) by the addition of a single nucleotide. Two adenines were found at positions 6817-18, instead of only one at 6817. The consequences are that the previously described ORFs 4 and 5 become one ORF (ORF 4). This ORF 4 would encode a predicted protein of 46.3 kDa, with a low homology of sequence with the inclusion body proteins of the caulimoviruses. De Kochko et al. (1998) also mention a possible ORF 5 (nts 7973-8136), which would encode a protein of 6.3 kDa with no apparent sequence homology with any protein in the sequence data bases.
The large intergenic region (736 nts long) contains an eucaryotic consensus TATA box and the promoter of transcription. A promoter fragment was isolated from CsVMV, comprising nucleotides -443 to +72. It was shown to direct strong constitutive gene expression in transgenic plants (Verdaguer et al., 1996). The functional architecture of the CsVMV promoter fragment has been reported (Verdaguer et al., 1998). The promoter is made up of different regions that confer distinct tissue-specific expression of the gene. For example, the region encompassing nucleotides -222 to -173 contains cis elements that control promoter expression in green tissues and root tips. The region encompassing nucleotides -178 to -63 is responsible for expression in vascular elements.
Relations with Cells and Tissues
Kitajima & Costa (1966) found spheroidal bodies, 50-60 µm in diameter, in ultra-thin sections by electron microscopy. CsVMV particles were found dispersed in certain areas of the cytoplasm that were rich in ribosomes but poor in other cytoplasmic organelles.
Ecology and Control
The impact of CsVMV on the culture of cassava is not well documented. A study showed that differences of yields between diseased and uninfected plants were observed but were not significant statistically (Santos et al., 1995). According to Calvert & Thresh (2002), a drought occurring at the beginning of the growth cycle would increase yield losses.
Figures
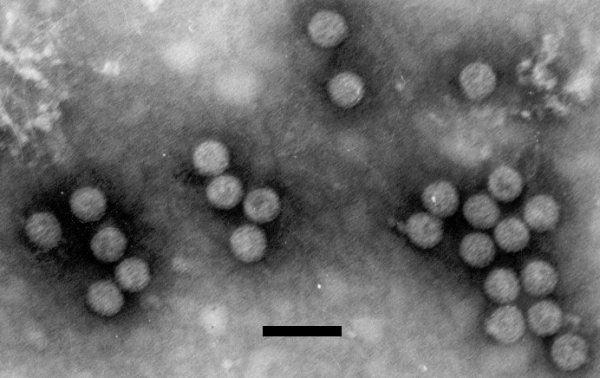
Electron micrograph of CsVMV particles. The bar represents 0.1 µm. Photograph courtesy of Pr E.W. Kitajima.
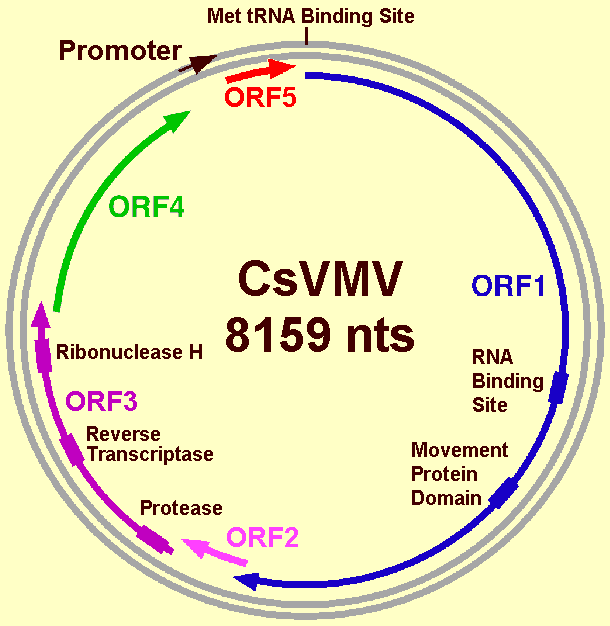
Predicted genomic organization of CsVMV according to the nucleotide sequence determined by de Kochko et al. (1998). The positions of the five open reading frames (ORFs) are indicated by arrows. Boxes represent conserved motifs of predicted proteins.
References list for DPV: Cassava vein mosaic virus (413)
- Calvert, Ospina & Shepherd, Journal of General Virology 76: 1271, 1995.
- Calvert & Thresh, in Cassava: Biology, Production and Utilization, p.239, ed R. J. Hillocks, J.M. Thresh, A.C. Bellotti, Wallingford: CABI Publishing, 2002.
- Costa, Jornal Agronomico 3: 239, 1940.
- De Kochko, Verdaguer, Taylor, Carcamo, Beachy & Fauquet, Archives of Virology 143: 945, 1998.
- Hull, Geering, Harper, Lockhart & Schoelz, in Virus Taxonomy, Classification and Nomenclature of Viruses, Eighth Report of the International Committee on the Taxonomy of Viruses, p 391, ed C.M. Fauquet, M.A. Mayo, J. Maniloff, U. Desselberger, L.A. Ball, London: Academic Press, 2005.
- Kitajima & Costa, Bragantia 25: 211, 1966.
- Lin & Kitajima, Fitopatologia Brasileira 5: 419, 1980.
- Santos, Gonçalves, Queiroz & Lima, Fitopatologia Brasileira 20: 506, 1995.
- Verdaguer, de Kochko, Beachy & Fauquet, Plant Molecular Biology 31: 1129, 1996.
- Verdaguer, de Kochko, Fux, Beachy & Fauquet, Plant Molecular Biology 37: 1055, 1998.

